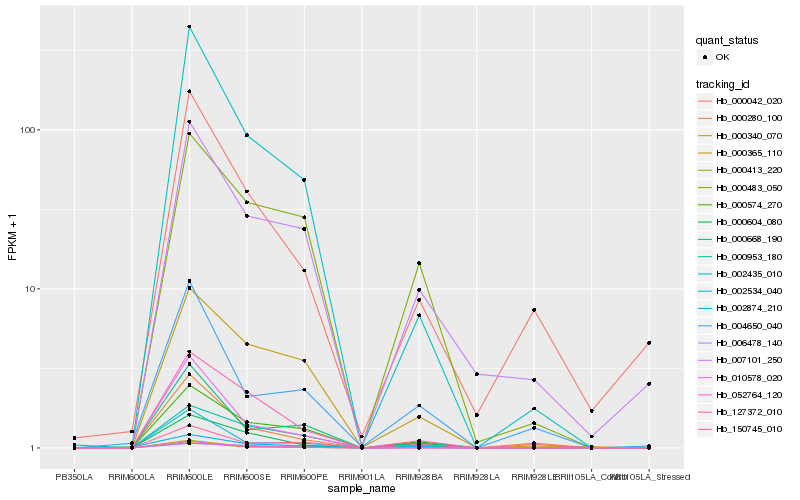
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_070 |
0.0 |
- |
- |
PREDICTED: CSC1-like protein At4g02900 [Jatropha curcas] |
| 2 |
Hb_002534_040 |
0.1096468077 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 3 |
Hb_000604_080 |
0.1223115748 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 4 |
Hb_002435_010 |
0.1236185311 |
- |
- |
PREDICTED: endochitinase PR4-like [Jatropha curcas] |
| 5 |
Hb_000574_270 |
0.1273712314 |
- |
- |
PREDICTED: VQ motif-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_150745_010 |
0.1321744201 |
- |
- |
hypothetical protein JCGZ_17337 [Jatropha curcas] |
| 7 |
Hb_000668_190 |
0.1358568971 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 8 |
Hb_000413_220 |
0.1497691605 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 9 |
Hb_002874_210 |
0.1505925342 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000280_100 |
0.1540299194 |
- |
- |
hypothetical protein JCGZ_13378 [Jatropha curcas] |
| 11 |
Hb_127372_010 |
0.1596849702 |
- |
- |
hypothetical protein POPTR_0017s06310g [Populus trichocarpa] |
| 12 |
Hb_004650_040 |
0.1619743546 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105638892 [Jatropha curcas] |
| 13 |
Hb_007101_250 |
0.1637461216 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 2 [Jatropha curcas] |
| 14 |
Hb_000042_020 |
0.1680581094 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_052764_120 |
0.1732621806 |
- |
- |
PREDICTED: uncharacterized protein LOC103941142 [Pyrus x bretschneideri] |
| 16 |
Hb_000953_180 |
0.1740063563 |
- |
- |
- |
| 17 |
Hb_000483_050 |
0.1765080995 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1-like [Jatropha curcas] |
| 18 |
Hb_006478_140 |
0.1785157173 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 19 |
Hb_010578_020 |
0.1788314886 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000365_110 |
0.1791334384 |
transcription factor |
TF Family: GRAS |
PREDICTED: protein SCARECROW [Jatropha curcas] |