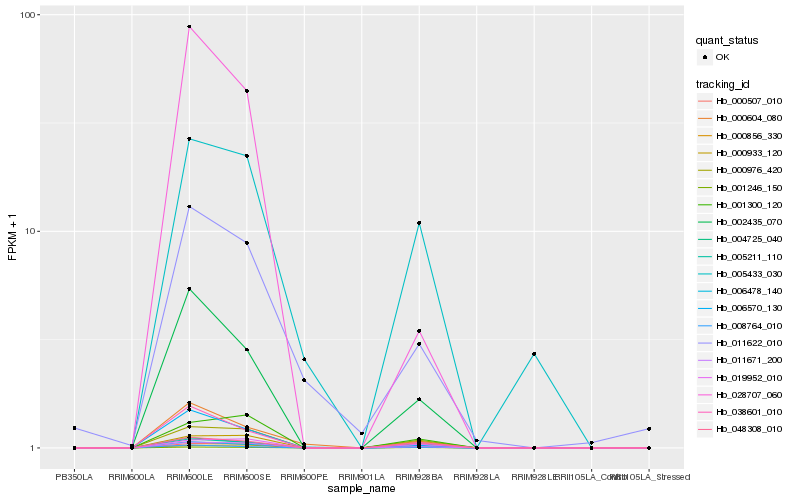
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004725_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_002435_070 |
0.0495630799 |
- |
- |
hypothetical protein JCGZ_05738 [Jatropha curcas] |
| 3 |
Hb_038601_010 |
0.0812423921 |
- |
- |
PREDICTED: uncharacterized protein LOC102606280 [Solanum tuberosum] |
| 4 |
Hb_011671_200 |
0.093837386 |
- |
- |
- |
| 5 |
Hb_006570_130 |
0.1075526734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_008764_010 |
0.1148031436 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000976_420 |
0.1219056706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_028707_060 |
0.1436997621 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC101308915 [Fragaria vesca subsp. vesca] |
| 9 |
Hb_005211_110 |
0.1559739558 |
- |
- |
hypothetical protein POPTR_0010s13790g [Populus trichocarpa] |
| 10 |
Hb_048308_010 |
0.1569298531 |
- |
- |
Peroxidase superfamily protein [Theobroma cacao] |
| 11 |
Hb_000604_080 |
0.1583629897 |
- |
- |
Fgenesh protein [Medicago truncatula] |
| 12 |
Hb_001246_150 |
0.1589334661 |
- |
- |
hypothetical protein MIMGU_mgv1a021562mg [Erythranthe guttata] |
| 13 |
Hb_000856_330 |
0.165493272 |
- |
- |
PREDICTED: protein QUIRKY isoform X1 [Cicer arietinum] |
| 14 |
Hb_000933_120 |
0.1711492344 |
- |
- |
hypothetical protein L484_011925 [Morus notabilis] |
| 15 |
Hb_019952_010 |
0.1815351668 |
- |
- |
- |
| 16 |
Hb_000507_010 |
0.182353767 |
- |
- |
hypothetical protein POPTR_0007s02420g [Populus trichocarpa] |
| 17 |
Hb_001300_120 |
0.184943723 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |
| 18 |
Hb_006478_140 |
0.1976328415 |
- |
- |
PREDICTED: vicilin-like antimicrobial peptides 2-2 [Jatropha curcas] |
| 19 |
Hb_011622_010 |
0.203666399 |
- |
- |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 20 |
Hb_005433_030 |
0.2114862343 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |