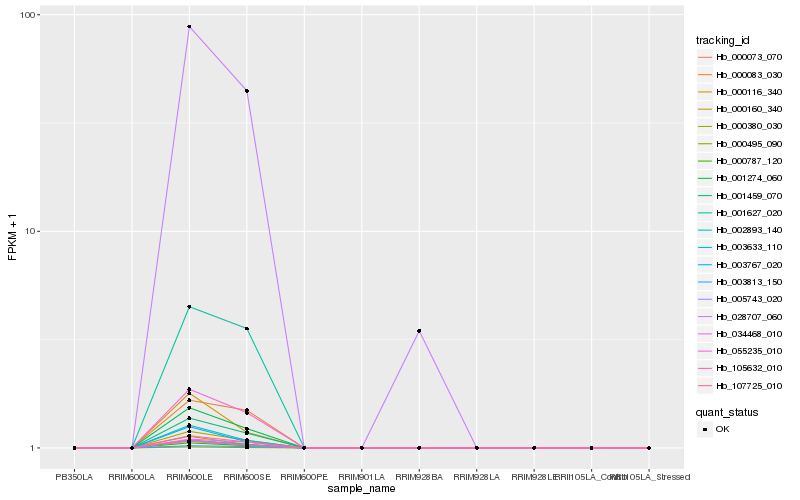
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000380_030 |
0.0 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 2 |
Hb_000787_120 |
0.0022842577 |
- |
- |
2S sulfur-rich seed storage protein precursor large subunit, putative [Ricinus communis] |
| 3 |
Hb_003633_110 |
0.003463964 |
- |
- |
hypothetical protein PRUPE_ppa019329mg [Prunus persica] |
| 4 |
Hb_001274_060 |
0.0048708501 |
- |
- |
PREDICTED: uncharacterized protein LOC105802656 [Gossypium raimondii] |
| 5 |
Hb_001459_070 |
0.0074603629 |
- |
- |
- |
| 6 |
Hb_107725_010 |
0.0127124762 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_055235_010 |
0.0346517131 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_005743_020 |
0.0364130156 |
- |
- |
- |
| 9 |
Hb_105632_010 |
0.036431326 |
- |
- |
- |
| 10 |
Hb_000495_090 |
0.0424641959 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 11 |
Hb_000073_070 |
0.0435407546 |
- |
- |
- |
| 12 |
Hb_002893_140 |
0.0692475557 |
- |
- |
Glycine-rich cell wall structural protein precursor, putative [Ricinus communis] |
| 13 |
Hb_003813_150 |
0.0736343515 |
- |
- |
hypothetical protein POPTR_0004s09690g [Populus trichocarpa] |
| 14 |
Hb_034468_010 |
0.0762163419 |
- |
- |
hypothetical protein EUGRSUZ_A02374 [Eucalyptus grandis] |
| 15 |
Hb_000160_340 |
0.0850093846 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_003767_020 |
0.0939432678 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |
| 17 |
Hb_028707_060 |
0.0998429692 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC101308915 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_001627_020 |
0.1039819521 |
- |
- |
- |
| 19 |
Hb_000083_030 |
0.1067629585 |
- |
- |
hypothetical protein JCGZ_24974 [Jatropha curcas] |
| 20 |
Hb_000116_340 |
0.1180981851 |
- |
- |
PREDICTED: small acidic protein 1 [Jatropha curcas] |