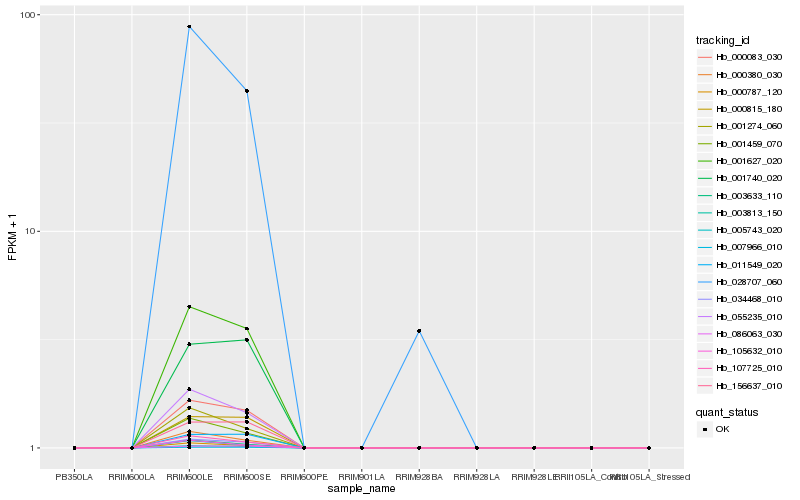
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003813_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0004s09690g [Populus trichocarpa] |
| 2 |
Hb_034468_010 |
0.0025878678 |
- |
- |
hypothetical protein EUGRSUZ_A02374 [Eucalyptus grandis] |
| 3 |
Hb_001627_020 |
0.0304398046 |
- |
- |
- |
| 4 |
Hb_000083_030 |
0.0332317354 |
- |
- |
hypothetical protein JCGZ_24974 [Jatropha curcas] |
| 5 |
Hb_055235_010 |
0.0390244515 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_107725_010 |
0.0609463396 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001459_070 |
0.0661896614 |
- |
- |
- |
| 8 |
Hb_003633_110 |
0.0701781337 |
- |
- |
hypothetical protein PRUPE_ppa019329mg [Prunus persica] |
| 9 |
Hb_000787_120 |
0.0713552938 |
- |
- |
2S sulfur-rich seed storage protein precursor large subunit, putative [Ricinus communis] |
| 10 |
Hb_000380_030 |
0.0736343515 |
- |
- |
Kiwellin, putative [Ricinus communis] |
| 11 |
Hb_001274_060 |
0.0784929161 |
- |
- |
PREDICTED: uncharacterized protein LOC105802656 [Gossypium raimondii] |
| 12 |
Hb_000815_180 |
0.0899448892 |
- |
- |
- |
| 13 |
Hb_156637_010 |
0.0975829659 |
- |
- |
PREDICTED: aldose reductase-like [Camelina sativa] |
| 14 |
Hb_011549_020 |
0.0986987881 |
- |
- |
- |
| 15 |
Hb_086063_030 |
0.1065830856 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8-like [Camelina sativa] |
| 16 |
Hb_007966_010 |
0.1072969396 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 17 |
Hb_028707_060 |
0.1080337069 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC101308915 [Fragaria vesca subsp. vesca] |
| 18 |
Hb_005743_020 |
0.1099144573 |
- |
- |
- |
| 19 |
Hb_105632_010 |
0.1099326762 |
- |
- |
- |
| 20 |
Hb_001740_020 |
0.1105643238 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |