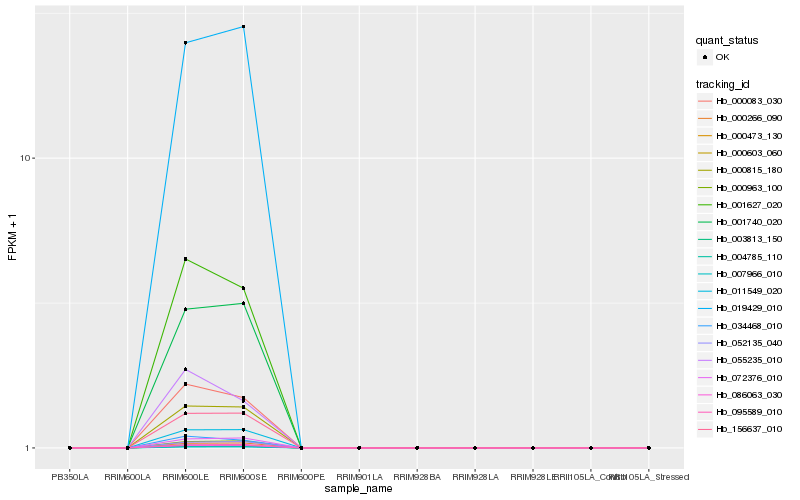
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000083_030 |
0.0 |
- |
- |
hypothetical protein JCGZ_24974 [Jatropha curcas] |
| 2 |
Hb_001627_020 |
0.002792972 |
- |
- |
- |
| 3 |
Hb_034468_010 |
0.0306448545 |
- |
- |
hypothetical protein EUGRSUZ_A02374 [Eucalyptus grandis] |
| 4 |
Hb_003813_150 |
0.0332317354 |
- |
- |
hypothetical protein POPTR_0004s09690g [Populus trichocarpa] |
| 5 |
Hb_000815_180 |
0.0567739614 |
- |
- |
- |
| 6 |
Hb_156637_010 |
0.0644258705 |
- |
- |
PREDICTED: aldose reductase-like [Camelina sativa] |
| 7 |
Hb_011549_020 |
0.0655438236 |
- |
- |
- |
| 8 |
Hb_055235_010 |
0.0722201482 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 9 |
Hb_086063_030 |
0.0734439766 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8-like [Camelina sativa] |
| 10 |
Hb_007966_010 |
0.0741593352 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 11 |
Hb_001740_020 |
0.0774337518 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 12 |
Hb_000266_090 |
0.0793589153 |
- |
- |
- |
| 13 |
Hb_004785_110 |
0.0853338853 |
- |
- |
- |
| 14 |
Hb_000603_060 |
0.0853748252 |
- |
- |
putative gag protein [Coffea canephora] |
| 15 |
Hb_000473_130 |
0.0853846497 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 16 |
Hb_095589_010 |
0.0855090925 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 17 |
Hb_052135_040 |
0.085776489 |
- |
- |
- |
| 18 |
Hb_000963_100 |
0.085837998 |
- |
- |
Protein SUA5, putative [Ricinus communis] |
| 19 |
Hb_072376_010 |
0.0861172502 |
- |
- |
- |
| 20 |
Hb_019429_010 |
0.0911394941 |
- |
- |
conserved hypothetical protein [Ricinus communis] |