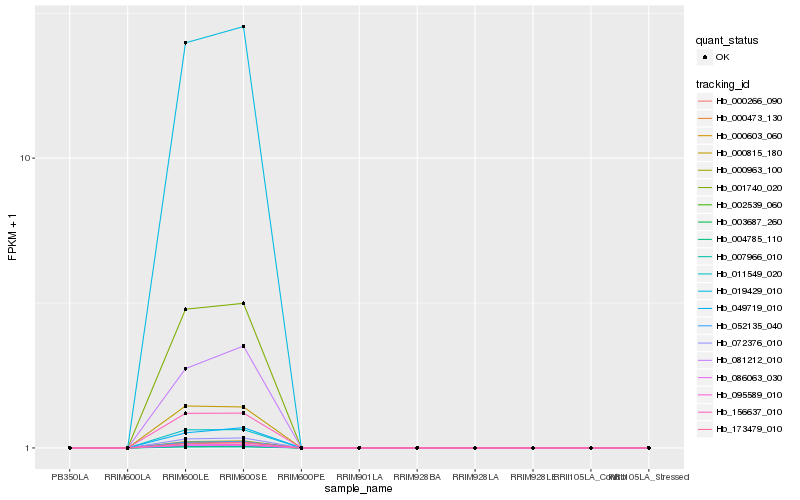
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086063_030 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8-like [Camelina sativa] |
| 2 |
Hb_007966_010 |
0.0007167143 |
- |
- |
carbohydrate binding protein, putative [Ricinus communis] |
| 3 |
Hb_001740_020 |
0.0039976688 |
- |
- |
hypothetical protein JCGZ_23765 [Jatropha curcas] |
| 4 |
Hb_000266_090 |
0.0059269307 |
- |
- |
- |
| 5 |
Hb_011549_020 |
0.0079133958 |
- |
- |
- |
| 6 |
Hb_156637_010 |
0.0090329666 |
- |
- |
PREDICTED: aldose reductase-like [Camelina sativa] |
| 7 |
Hb_004785_110 |
0.0119158213 |
- |
- |
- |
| 8 |
Hb_000603_060 |
0.0119568629 |
- |
- |
putative gag protein [Coffea canephora] |
| 9 |
Hb_000473_130 |
0.0119667118 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 10 |
Hb_095589_010 |
0.0120914642 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 11 |
Hb_052135_040 |
0.0123595285 |
- |
- |
- |
| 12 |
Hb_000963_100 |
0.0124211916 |
- |
- |
Protein SUA5, putative [Ricinus communis] |
| 13 |
Hb_072376_010 |
0.012701146 |
- |
- |
- |
| 14 |
Hb_000815_180 |
0.0166942423 |
- |
- |
- |
| 15 |
Hb_019429_010 |
0.0177366974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_173479_010 |
0.0387807216 |
- |
- |
- |
| 17 |
Hb_003687_260 |
0.051077081 |
- |
- |
PREDICTED: inorganic phosphate transporter 1-4-like [Camelina sativa] |
| 18 |
Hb_002539_060 |
0.0520428779 |
- |
- |
- |
| 19 |
Hb_049719_010 |
0.0560826353 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_081212_010 |
0.0633819219 |
- |
- |
- |