| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
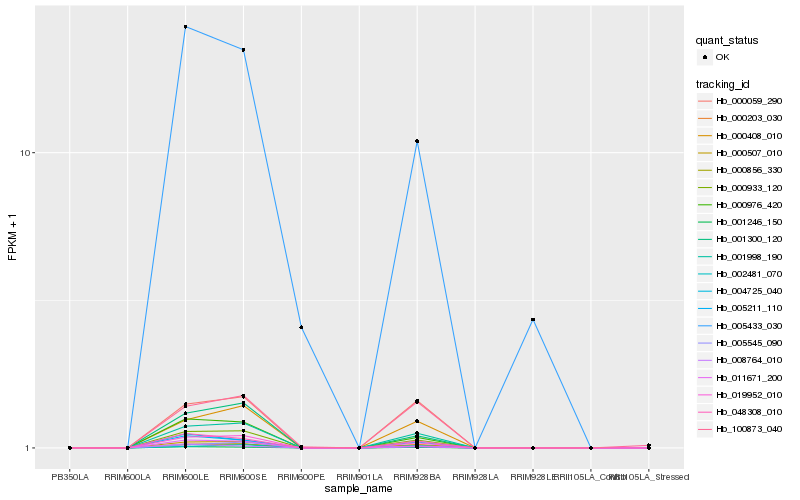
Hb_000933_120 |
0.0 |
- |
- |
hypothetical protein L484_011925 [Morus notabilis] |
| 2 |
Hb_000507_010 |
0.0122041988 |
- |
- |
hypothetical protein POPTR_0007s02420g [Populus trichocarpa] |
| 3 |
Hb_019952_010 |
0.0198533724 |
- |
- |
- |
| 4 |
Hb_000976_420 |
0.0504273969 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001998_190 |
0.054731951 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 6 |
Hb_000856_330 |
0.0788012951 |
- |
- |
PREDICTED: protein QUIRKY isoform X1 [Cicer arietinum] |
| 7 |
Hb_001300_120 |
0.0915513026 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |
| 8 |
Hb_001246_150 |
0.0940234218 |
- |
- |
hypothetical protein MIMGU_mgv1a021562mg [Erythranthe guttata] |
| 9 |
Hb_011671_200 |
0.0964095361 |
- |
- |
- |
| 10 |
Hb_005211_110 |
0.1035000642 |
- |
- |
hypothetical protein POPTR_0010s13790g [Populus trichocarpa] |
| 11 |
Hb_048308_010 |
0.1041058797 |
- |
- |
Peroxidase superfamily protein [Theobroma cacao] |
| 12 |
Hb_000408_010 |
0.1152135192 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 13 |
Hb_005545_090 |
0.1224039012 |
- |
- |
- |
| 14 |
Hb_000203_030 |
0.1247715519 |
- |
- |
- |
| 15 |
Hb_002481_070 |
0.146196635 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 16 |
Hb_000059_290 |
0.1464928748 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 17 |
Hb_008764_010 |
0.1659219961 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004725_040 |
0.1711492344 |
- |
- |
- |
| 19 |
Hb_005433_030 |
0.1730196642 |
transcription factor |
TF Family: MYB |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_100873_040 |
0.178644455 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |