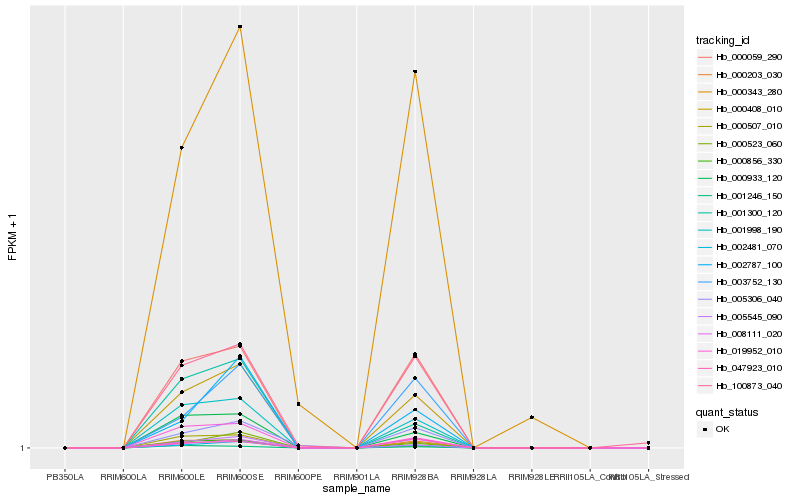
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000408_010 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 2 |
Hb_001998_190 |
0.066682522 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 3 |
Hb_002481_070 |
0.0683117131 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 4 |
Hb_000059_290 |
0.0774133385 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 5 |
Hb_000203_030 |
0.0849864993 |
- |
- |
- |
| 6 |
Hb_000507_010 |
0.1050465969 |
- |
- |
hypothetical protein POPTR_0007s02420g [Populus trichocarpa] |
| 7 |
Hb_019952_010 |
0.1099774707 |
- |
- |
- |
| 8 |
Hb_000933_120 |
0.1152135192 |
- |
- |
hypothetical protein L484_011925 [Morus notabilis] |
| 9 |
Hb_100873_040 |
0.1222642629 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 10 |
Hb_008111_020 |
0.1351342576 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |
| 11 |
Hb_005545_090 |
0.1353336212 |
- |
- |
- |
| 12 |
Hb_003752_130 |
0.1444985099 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 13 |
Hb_000343_280 |
0.1457717919 |
- |
- |
hypothetical protein JCGZ_24094 [Jatropha curcas] |
| 14 |
Hb_005306_040 |
0.151708474 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_002787_100 |
0.151762626 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 16 |
Hb_000523_060 |
0.1520973335 |
- |
- |
- |
| 17 |
Hb_001300_120 |
0.1636950974 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |
| 18 |
Hb_047923_010 |
0.164797919 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 19 |
Hb_001246_150 |
0.1650835603 |
- |
- |
hypothetical protein MIMGU_mgv1a021562mg [Erythranthe guttata] |
| 20 |
Hb_000856_330 |
0.1650846089 |
- |
- |
PREDICTED: protein QUIRKY isoform X1 [Cicer arietinum] |