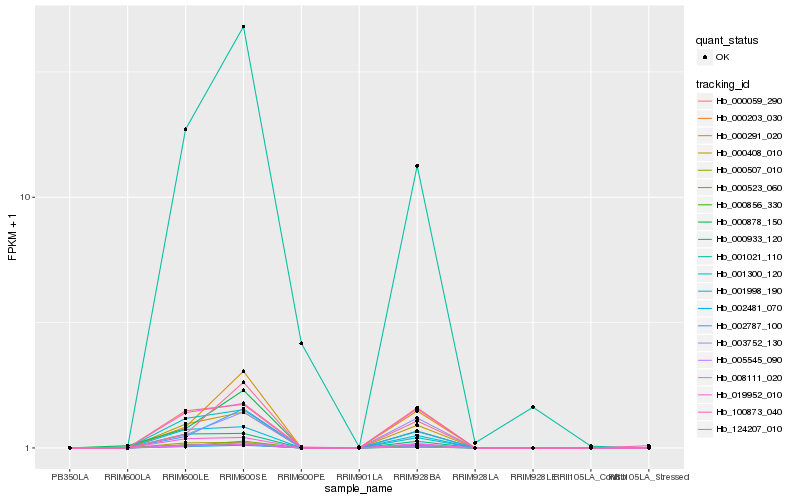
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002481_070 |
0.0 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 2 |
Hb_000408_010 |
0.0683117131 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 3 |
Hb_000523_060 |
0.0841040123 |
- |
- |
- |
| 4 |
Hb_002787_100 |
0.0911367237 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 5 |
Hb_005545_090 |
0.0997773204 |
- |
- |
- |
| 6 |
Hb_001998_190 |
0.1163448523 |
transcription factor |
TF Family: CSD |
PREDICTED: uncharacterized protein LOC105636942 [Jatropha curcas] |
| 7 |
Hb_000291_020 |
0.1223198352 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 8 |
Hb_019952_010 |
0.1324192656 |
- |
- |
- |
| 9 |
Hb_000507_010 |
0.1341324851 |
- |
- |
hypothetical protein POPTR_0007s02420g [Populus trichocarpa] |
| 10 |
Hb_001021_110 |
0.1419155158 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 11 |
Hb_000059_290 |
0.1431471688 |
- |
- |
pterin-4-alpha-carbinolamine dehydratase, putative [Ricinus communis] |
| 12 |
Hb_003752_130 |
0.1461127628 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 13 |
Hb_000933_120 |
0.146196635 |
- |
- |
hypothetical protein L484_011925 [Morus notabilis] |
| 14 |
Hb_000878_150 |
0.1492750068 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 15 |
Hb_124207_010 |
0.1528836746 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_000203_030 |
0.1529712638 |
- |
- |
- |
| 17 |
Hb_001300_120 |
0.1547937484 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |
| 18 |
Hb_000856_330 |
0.1646025599 |
- |
- |
PREDICTED: protein QUIRKY isoform X1 [Cicer arietinum] |
| 19 |
Hb_100873_040 |
0.1651857124 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 20 |
Hb_008111_020 |
0.16584816 |
- |
- |
Receptor like protein 6, putative [Theobroma cacao] |