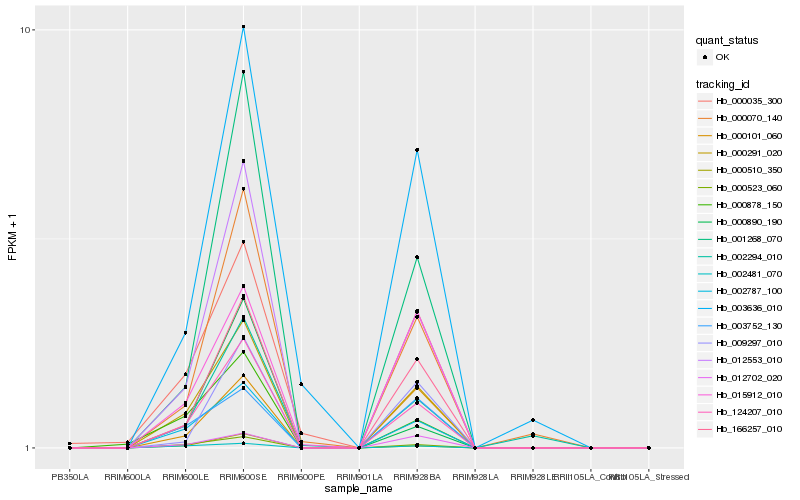
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_124207_010 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_000291_020 |
0.0376060034 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 3 |
Hb_012553_010 |
0.0588218211 |
- |
- |
hypothetical protein JCGZ_10552 [Jatropha curcas] |
| 4 |
Hb_002787_100 |
0.0624604506 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 5 |
Hb_166257_010 |
0.0941021396 |
- |
- |
hypothetical protein JCGZ_21808 [Jatropha curcas] |
| 6 |
Hb_000523_060 |
0.0988540491 |
- |
- |
- |
| 7 |
Hb_001268_070 |
0.1140319851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000510_350 |
0.1312317962 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 9 |
Hb_000070_140 |
0.1367287906 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 10 |
Hb_000878_150 |
0.1439818498 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 11 |
Hb_002481_070 |
0.1528836746 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 12 |
Hb_000035_300 |
0.1612664633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_015912_010 |
0.1659235903 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_009297_010 |
0.1661200429 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_003636_010 |
0.1667069472 |
- |
- |
hypothetical protein CISIN_1g037850mg, partial [Citrus sinensis] |
| 16 |
Hb_012702_020 |
0.1685791643 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 17 |
Hb_002294_010 |
0.1691639986 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 18 |
Hb_003752_130 |
0.1709191171 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F1-like [Jatropha curcas] |
| 19 |
Hb_000101_060 |
0.171878022 |
- |
- |
- |
| 20 |
Hb_000890_190 |
0.1807640339 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |