| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
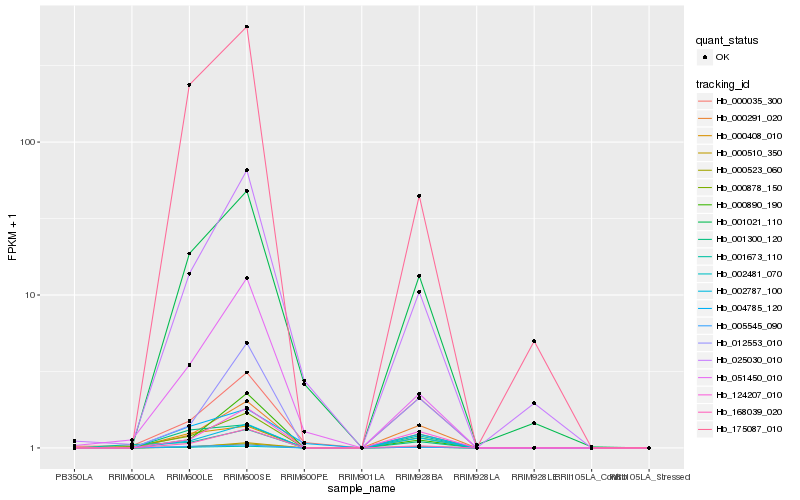
Hb_000878_150 |
0.0 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 2 |
Hb_000523_060 |
0.1036779266 |
- |
- |
- |
| 3 |
Hb_002787_100 |
0.125891614 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 4 |
Hb_000291_020 |
0.1428778443 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 5 |
Hb_124207_010 |
0.1439818498 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 6 |
Hb_000510_350 |
0.1456238865 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 7 |
Hb_002481_070 |
0.1492750068 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 8 |
Hb_005545_090 |
0.1509686151 |
- |
- |
- |
| 9 |
Hb_051450_010 |
0.1540853816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_168039_020 |
0.1568153188 |
- |
- |
- |
| 11 |
Hb_001021_110 |
0.1595602054 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 12 |
Hb_025030_010 |
0.1731776791 |
- |
- |
- |
| 13 |
Hb_012553_010 |
0.1734930443 |
- |
- |
hypothetical protein JCGZ_10552 [Jatropha curcas] |
| 14 |
Hb_000035_300 |
0.1922341167 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000890_190 |
0.1950699503 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 16 |
Hb_175087_010 |
0.199408493 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 17 |
Hb_000408_010 |
0.2043038062 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 18 |
Hb_004785_120 |
0.2044517831 |
- |
- |
Receptor protein kinase 1 [Theobroma cacao] |
| 19 |
Hb_001673_110 |
0.2048201443 |
- |
- |
PREDICTED: RING-H2 finger protein ATL70-like [Pyrus x bretschneideri] |
| 20 |
Hb_001300_120 |
0.207910017 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |