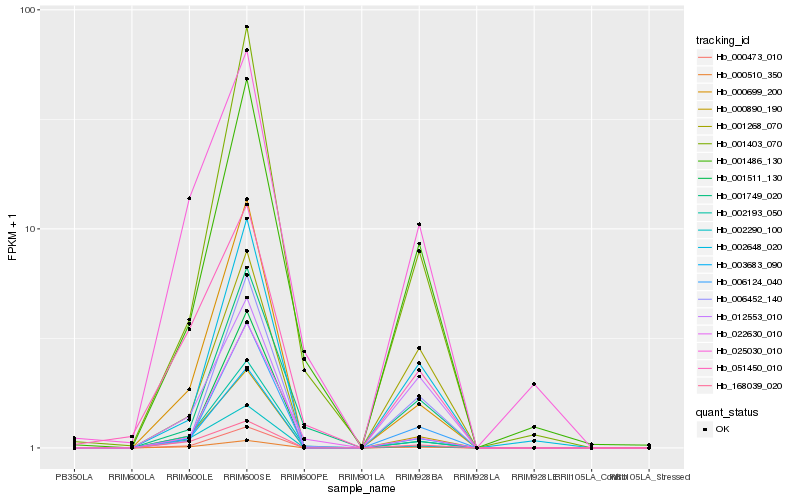
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_190 |
0.0 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 2 |
Hb_003683_090 |
0.0403861255 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 3 |
Hb_000510_350 |
0.0626066739 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 4 |
Hb_000699_200 |
0.0865290596 |
- |
- |
hypothetical protein POPTR_0019s02340g [Populus trichocarpa] |
| 5 |
Hb_000473_010 |
0.0878116635 |
- |
- |
PREDICTED: galactolipase DONGLE, chloroplastic [Jatropha curcas] |
| 6 |
Hb_168039_020 |
0.0949226457 |
- |
- |
- |
| 7 |
Hb_002193_050 |
0.1061447789 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 8 |
Hb_006124_040 |
0.1171877878 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 9 |
Hb_002290_100 |
0.1270984719 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 10 |
Hb_002648_020 |
0.1293297293 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Populus euphratica] |
| 11 |
Hb_001403_070 |
0.137400326 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X1 [Populus euphratica] |
| 12 |
Hb_051450_010 |
0.1509487917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012553_010 |
0.1539628411 |
- |
- |
hypothetical protein JCGZ_10552 [Jatropha curcas] |
| 14 |
Hb_001486_130 |
0.1560796716 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB5-like [Jatropha curcas] |
| 15 |
Hb_025030_010 |
0.158586116 |
- |
- |
- |
| 16 |
Hb_001268_070 |
0.1590296367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_006452_140 |
0.1599280833 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 18 |
Hb_001749_020 |
0.1679858735 |
- |
- |
hypothetical protein CISIN_1g042220mg, partial [Citrus sinensis] |
| 19 |
Hb_001511_130 |
0.1714775428 |
- |
- |
PREDICTED: taxadien-5-alpha-ol O-acetyltransferase [Jatropha curcas] |
| 20 |
Hb_022630_010 |
0.1758058221 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |