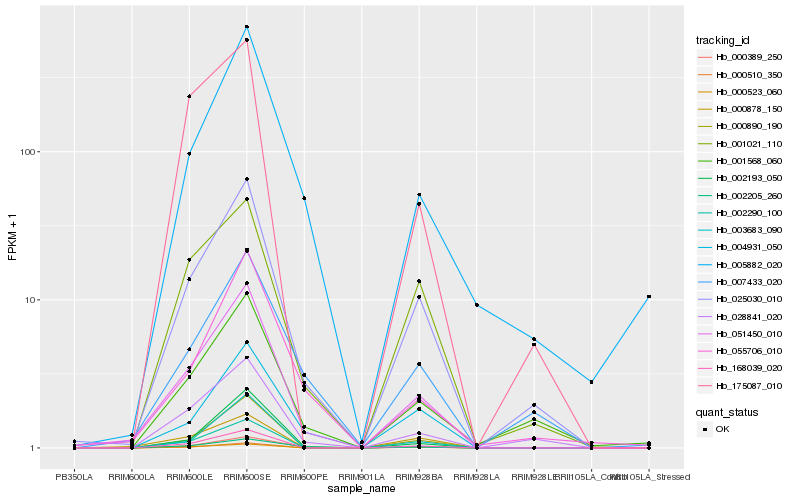
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_051450_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_025030_010 |
0.0999257193 |
- |
- |
- |
| 3 |
Hb_168039_020 |
0.1186083545 |
- |
- |
- |
| 4 |
Hb_000510_350 |
0.1347428906 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 5 |
Hb_000389_250 |
0.1414225889 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21556 [Jatropha curcas] |
| 6 |
Hb_000890_190 |
0.1509487917 |
- |
- |
glutaredoxin, grx, putative [Ricinus communis] |
| 7 |
Hb_002193_050 |
0.1525190592 |
- |
- |
PREDICTED: cytochrome P450 93A1-like [Citrus sinensis] |
| 8 |
Hb_002290_100 |
0.1525394312 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 9 |
Hb_002205_260 |
0.1528562402 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 10 |
Hb_000878_150 |
0.1540853816 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 11 |
Hb_028841_020 |
0.1577720433 |
- |
- |
hypothetical protein CISIN_1g041345mg [Citrus sinensis] |
| 12 |
Hb_004931_050 |
0.1597361117 |
transcription factor |
TF Family: MYB-related |
Transcription repressor MYB4, putative [Ricinus communis] |
| 13 |
Hb_055706_010 |
0.1610066816 |
- |
- |
hypothetical protein JCGZ_22222 [Jatropha curcas] |
| 14 |
Hb_001021_110 |
0.1673005363 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 15 |
Hb_005882_020 |
0.1703617057 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 16 |
Hb_007433_020 |
0.1733677801 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 17 |
Hb_003683_090 |
0.1740597447 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 18 |
Hb_001568_060 |
0.1782837438 |
- |
- |
PREDICTED: uncharacterized protein LOC105650728 [Jatropha curcas] |
| 19 |
Hb_175087_010 |
0.1798147731 |
- |
- |
hypothetical protein POPTR_0011s16980g, partial [Populus trichocarpa] |
| 20 |
Hb_000523_060 |
0.1836678536 |
- |
- |
- |