| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
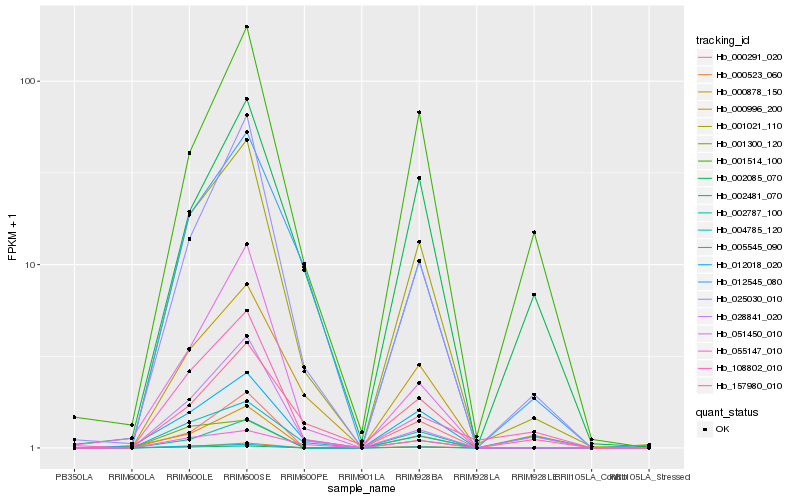
Hb_001021_110 |
0.0 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 2 |
Hb_004785_120 |
0.0947858231 |
- |
- |
Receptor protein kinase 1 [Theobroma cacao] |
| 3 |
Hb_000523_060 |
0.1255396212 |
- |
- |
- |
| 4 |
Hb_025030_010 |
0.1263820922 |
- |
- |
- |
| 5 |
Hb_000996_200 |
0.129710307 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Populus euphratica] |
| 6 |
Hb_005545_090 |
0.1335016204 |
- |
- |
- |
| 7 |
Hb_002481_070 |
0.1419155158 |
- |
- |
hypothetical protein JCGZ_18787 [Jatropha curcas] |
| 8 |
Hb_002787_100 |
0.1517757677 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 9 |
Hb_012545_080 |
0.1581329783 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 10 |
Hb_055147_010 |
0.1583722326 |
- |
- |
thioredoxin H-type 7 [Hevea brasiliensis] |
| 11 |
Hb_000878_150 |
0.1595602054 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 12 |
Hb_012018_020 |
0.1622720305 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 13 |
Hb_001514_100 |
0.1630648896 |
- |
- |
PREDICTED: F-box protein GID2 [Jatropha curcas] |
| 14 |
Hb_157980_010 |
0.1644352576 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_108802_010 |
0.1669737743 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 16 |
Hb_051450_010 |
0.1673005363 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_028841_020 |
0.170178387 |
- |
- |
hypothetical protein CISIN_1g041345mg [Citrus sinensis] |
| 18 |
Hb_002085_070 |
0.1745376233 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_000291_020 |
0.1747646993 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 20 |
Hb_001300_120 |
0.1782253639 |
- |
- |
PREDICTED: stress response protein nst1-like [Jatropha curcas] |