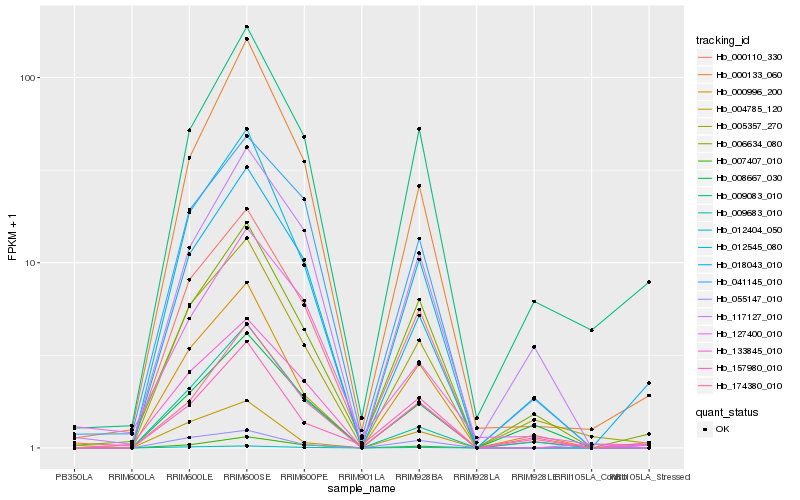
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_330 |
0.0 |
- |
- |
PREDICTED: probable nitrite transporter At1g68570-like [Glycine max] |
| 2 |
Hb_012404_050 |
0.0827116933 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 3 |
Hb_041145_010 |
0.090401896 |
- |
- |
hypothetical protein CISIN_1g021653mg [Citrus sinensis] |
| 4 |
Hb_012545_080 |
0.0932759379 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 5 |
Hb_007407_010 |
0.0998569704 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 74 [Populus euphratica] |
| 6 |
Hb_005357_270 |
0.1075062291 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_000133_060 |
0.110665821 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_133845_010 |
0.1138250346 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_006634_080 |
0.1173821545 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 10 |
Hb_000996_200 |
0.1214399795 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Populus euphratica] |
| 11 |
Hb_055147_010 |
0.1289661167 |
- |
- |
thioredoxin H-type 7 [Hevea brasiliensis] |
| 12 |
Hb_117127_010 |
0.133413793 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH106 [Jatropha curcas] |
| 13 |
Hb_009683_010 |
0.1454109025 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 14 |
Hb_018043_010 |
0.147096242 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 15 |
Hb_174380_010 |
0.1490842052 |
- |
- |
kinase, putative [Ricinus communis] |
| 16 |
Hb_009083_010 |
0.1505953492 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 17 |
Hb_127400_010 |
0.1509895536 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 18 |
Hb_157980_010 |
0.152517521 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 19 |
Hb_004785_120 |
0.1546493789 |
- |
- |
Receptor protein kinase 1 [Theobroma cacao] |
| 20 |
Hb_008667_030 |
0.1568660444 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |