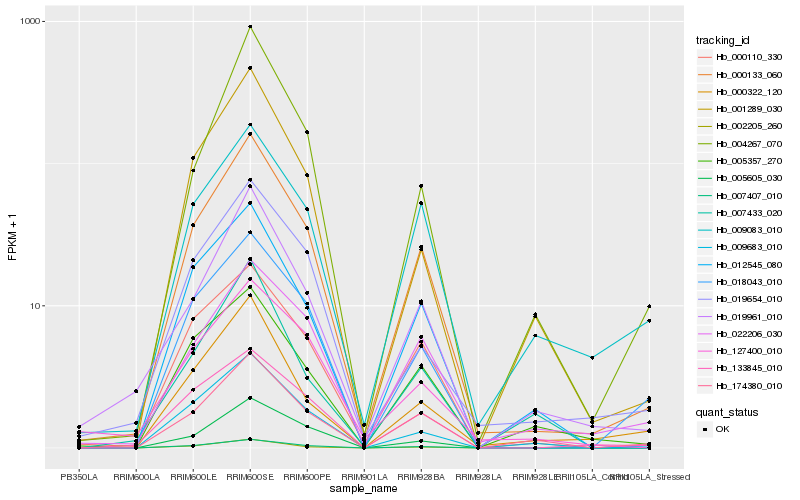
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000133_060 |
0.0 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 2 |
Hb_007407_010 |
0.0771566368 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 74 [Populus euphratica] |
| 3 |
Hb_002205_260 |
0.1065593192 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 55 [Jatropha curcas] |
| 4 |
Hb_000110_330 |
0.110665821 |
- |
- |
PREDICTED: probable nitrite transporter At1g68570-like [Glycine max] |
| 5 |
Hb_012545_080 |
0.111575161 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 6 |
Hb_019961_010 |
0.1149335098 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 7 |
Hb_174380_010 |
0.1161277981 |
- |
- |
kinase, putative [Ricinus communis] |
| 8 |
Hb_018043_010 |
0.1164241514 |
- |
- |
PREDICTED: auxin-induced protein 15A-like [Jatropha curcas] |
| 9 |
Hb_133845_010 |
0.1198324294 |
transcription factor |
TF Family: NAC |
transcription factor, putative [Ricinus communis] |
| 10 |
Hb_005605_030 |
0.1225612362 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 11 |
Hb_009683_010 |
0.1266299989 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_022206_030 |
0.1280887099 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 13 |
Hb_001289_030 |
0.1321806835 |
- |
- |
syntaxin, plant, putative [Ricinus communis] |
| 14 |
Hb_004267_070 |
0.1353790978 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 15 |
Hb_005357_270 |
0.135576334 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_019654_010 |
0.1355777041 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 17 |
Hb_009083_010 |
0.1358936456 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 18 |
Hb_000322_120 |
0.1424294873 |
- |
- |
PREDICTED: uncharacterized protein LOC105637900 [Jatropha curcas] |
| 19 |
Hb_007433_020 |
0.1426671712 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 20 |
Hb_127400_010 |
0.1427378437 |
desease resistance |
Gene Name: NB-ARC |
phosphoprotein phosphatase, putative [Ricinus communis] |