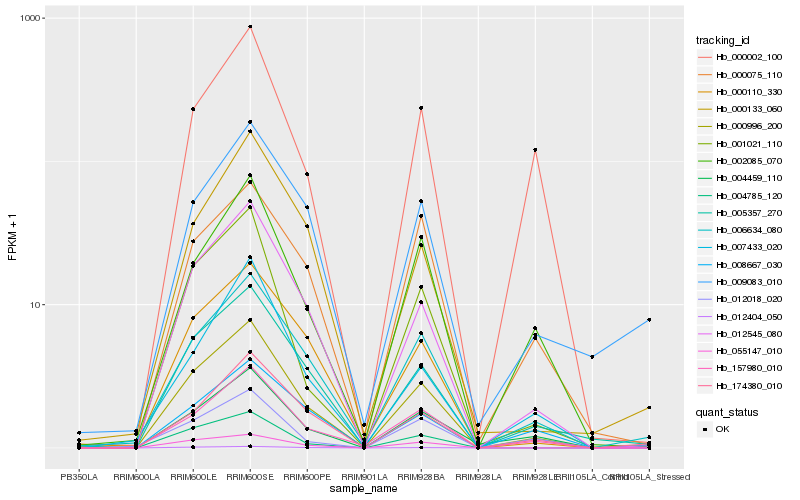
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000996_200 |
0.0 |
- |
- |
PREDICTED: (+)-neomenthol dehydrogenase-like [Populus euphratica] |
| 2 |
Hb_012545_080 |
0.0726093373 |
- |
- |
PREDICTED: uncharacterized protein LOC105633680 [Jatropha curcas] |
| 3 |
Hb_005357_270 |
0.0901329706 |
- |
- |
Glucose-6-phosphate/phosphate translocator 2, chloroplast precursor, putative [Ricinus communis] |
| 4 |
Hb_006634_080 |
0.0933481717 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-3b [Jatropha curcas] |
| 5 |
Hb_157980_010 |
0.1018415532 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 6 |
Hb_004459_110 |
0.1159385087 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 7 |
Hb_004785_120 |
0.1188052129 |
- |
- |
Receptor protein kinase 1 [Theobroma cacao] |
| 8 |
Hb_174380_010 |
0.1198687319 |
- |
- |
kinase, putative [Ricinus communis] |
| 9 |
Hb_000110_330 |
0.1214399795 |
- |
- |
PREDICTED: probable nitrite transporter At1g68570-like [Glycine max] |
| 10 |
Hb_055147_010 |
0.1269728591 |
- |
- |
thioredoxin H-type 7 [Hevea brasiliensis] |
| 11 |
Hb_002085_070 |
0.1281575728 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 12 |
Hb_001021_110 |
0.129710307 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 13 |
Hb_012404_050 |
0.1375690065 |
- |
- |
PREDICTED: uncharacterized protein LOC101496717 [Cicer arietinum] |
| 14 |
Hb_009083_010 |
0.1422519535 |
- |
- |
Phytosulfokines 3 precursor family protein [Populus trichocarpa] |
| 15 |
Hb_007433_020 |
0.14496592 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |
| 16 |
Hb_000133_060 |
0.147414028 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 17 |
Hb_000075_110 |
0.1485803979 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 18 |
Hb_008667_030 |
0.148751464 |
- |
- |
PREDICTED: premnaspirodiene oxygenase-like [Jatropha curcas] |
| 19 |
Hb_012018_020 |
0.1518269604 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Eucalyptus grandis] |
| 20 |
Hb_000002_100 |
0.1534069689 |
- |
- |
phosphate transporter [Manihot esculenta] |