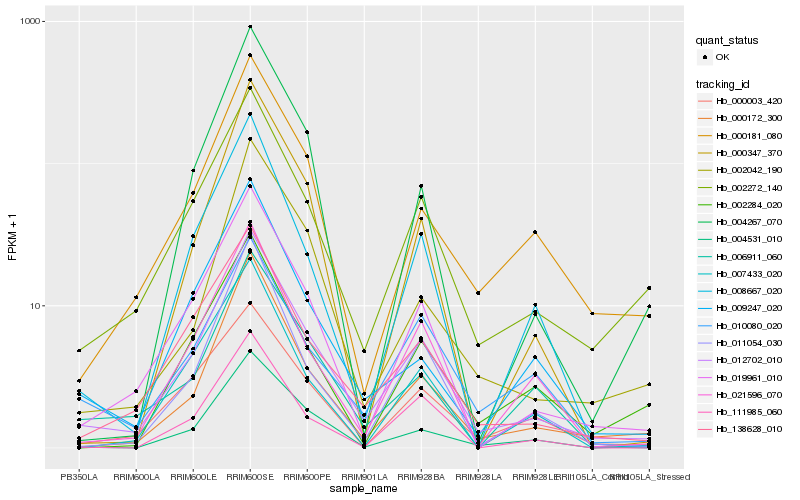
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012702_010 |
0.0 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 2 |
Hb_019961_010 |
0.098638637 |
- |
- |
hypothetical protein POPTR_0017s11350g [Populus trichocarpa] |
| 3 |
Hb_002272_140 |
0.1038596823 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 4 |
Hb_138628_010 |
0.1056412374 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 5 |
Hb_000172_300 |
0.11203182 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 6 |
Hb_004531_010 |
0.1260672812 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 7 |
Hb_011054_030 |
0.1300988848 |
- |
- |
PREDICTED: uncharacterized protein LOC103324687 [Prunus mume] |
| 8 |
Hb_021596_070 |
0.1306425196 |
rubber biosynthesis |
Gene Name: 1-deoxy-D-xylulose-5-phosphate synthase |
1-deoxy-D-xylulose 5-phosphate synthase [Hevea brasiliensis] |
| 9 |
Hb_008667_020 |
0.1323386785 |
- |
- |
PREDICTED: uncharacterized protein LOC101299609 [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000181_080 |
0.1327884514 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 11 |
Hb_010080_020 |
0.1411204835 |
- |
- |
PREDICTED: uncharacterized protein LOC105635249 [Jatropha curcas] |
| 12 |
Hb_000003_420 |
0.1426072945 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 13 |
Hb_006911_060 |
0.1442484922 |
- |
- |
hypothetical protein CISIN_1g000594mg [Citrus sinensis] |
| 14 |
Hb_009247_020 |
0.144806914 |
- |
- |
hypothetical protein JCGZ_04985 [Jatropha curcas] |
| 15 |
Hb_002042_190 |
0.1457708815 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 16 |
Hb_000347_370 |
0.1469376378 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 17 |
Hb_111985_060 |
0.1492950267 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 18 |
Hb_002284_020 |
0.1496294238 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 19 |
Hb_004267_070 |
0.1503548375 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 20 |
Hb_007433_020 |
0.1506018281 |
- |
- |
hypothetical protein VITISV_005320 [Vitis vinifera] |