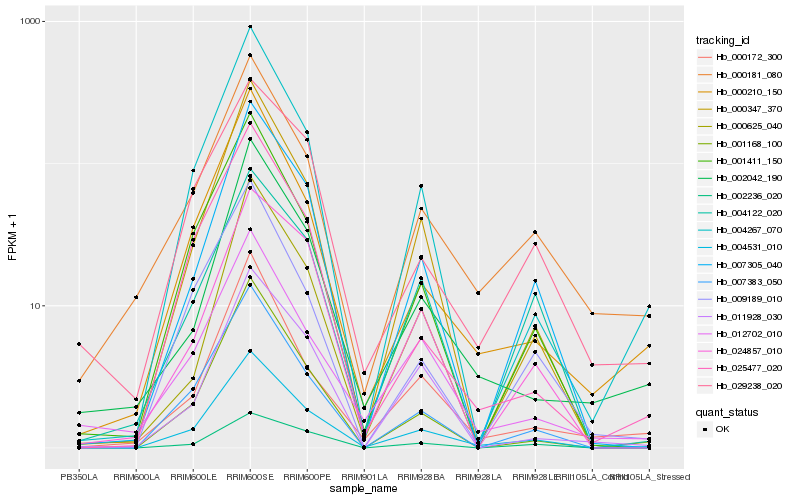
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004531_010 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |
| 2 |
Hb_000347_370 |
0.106743018 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 3 |
Hb_004267_070 |
0.1068357186 |
- |
- |
latex allene oxide synthase [Hevea brasiliensis] |
| 4 |
Hb_007305_040 |
0.1070616513 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 5 |
Hb_007383_050 |
0.108134608 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF061 [Jatropha curcas] |
| 6 |
Hb_001411_150 |
0.1111664454 |
- |
- |
Dormancy/auxin associated family protein, putative [Theobroma cacao] |
| 7 |
Hb_000210_150 |
0.1140162514 |
- |
- |
PREDICTED: protein YLS9-like [Jatropha curcas] |
| 8 |
Hb_000181_080 |
0.116745464 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 9 |
Hb_012702_010 |
0.1260672812 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X4 [Vitis vinifera] |
| 10 |
Hb_001168_100 |
0.1266381621 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FLS2-like [Citrus sinensis] |
| 11 |
Hb_002042_190 |
0.1298003142 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase ATL6-like [Jatropha curcas] |
| 12 |
Hb_000172_300 |
0.1305530344 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 13 |
Hb_029238_020 |
0.134928638 |
- |
- |
PREDICTED: expansin-like A1 [Jatropha curcas] |
| 14 |
Hb_009189_010 |
0.1382658466 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 15 |
Hb_025477_020 |
0.1405828676 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF027 [Jatropha curcas] |
| 16 |
Hb_024857_010 |
0.141408613 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 17 |
Hb_002236_020 |
0.1427723292 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 18 |
Hb_011928_030 |
0.1429953628 |
- |
- |
PREDICTED: probable calcium-binding protein CML23 [Jatropha curcas] |
| 19 |
Hb_000625_040 |
0.1447561325 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 20 |
Hb_004122_020 |
0.1451635432 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |