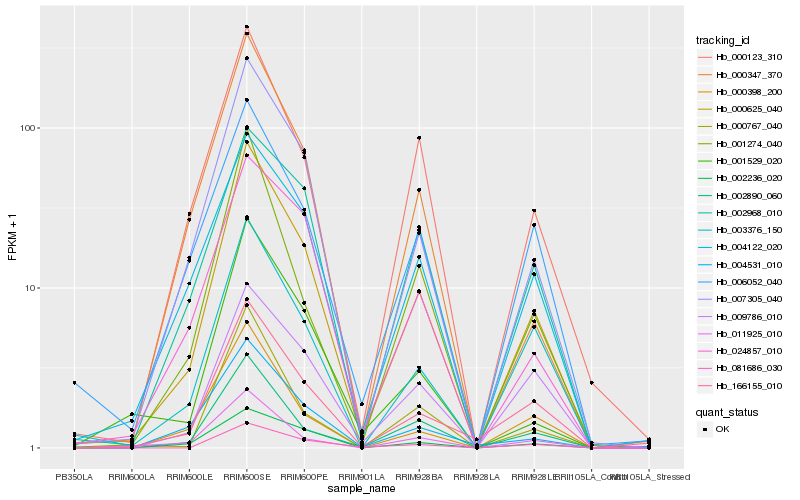
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000625_040 |
0.0 |
- |
- |
PREDICTED: pathogenesis-related protein 5-like [Jatropha curcas] |
| 2 |
Hb_007305_040 |
0.0776995432 |
- |
- |
Nodulin-26, putative [Ricinus communis] |
| 3 |
Hb_003376_150 |
0.1147950303 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 4 |
Hb_166155_010 |
0.1148097628 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 5 |
Hb_002890_060 |
0.121395373 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 6 |
Hb_000123_310 |
0.127884389 |
- |
- |
PREDICTED: aquaporin TIP1-3 [Jatropha curcas] |
| 7 |
Hb_004122_020 |
0.1280515479 |
transcription factor |
TF Family: ERF |
ERF/AP2 domain-containing transcription factor [Manihot esculenta] |
| 8 |
Hb_002236_020 |
0.1310068184 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 9 |
Hb_009786_010 |
0.1320052292 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 10 |
Hb_000398_200 |
0.1358135379 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 11 |
Hb_000347_370 |
0.1364667843 |
- |
- |
PREDICTED: cytochrome P450 71A1-like [Jatropha curcas] |
| 12 |
Hb_024857_010 |
0.1377059836 |
- |
- |
glutaredoxin 2.1 [Hevea brasiliensis] |
| 13 |
Hb_006052_040 |
0.1377982086 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 14 |
Hb_011925_010 |
0.1389655273 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_081686_030 |
0.1399893634 |
- |
- |
hypothetical protein MIMGU_mgv11b016770mg [Erythranthe guttata] |
| 16 |
Hb_001529_020 |
0.141435383 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000767_040 |
0.1431321915 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_001274_040 |
0.1432324809 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 19 |
Hb_002968_010 |
0.1445703005 |
- |
- |
PREDICTED: pectinesterase [Jatropha curcas] |
| 20 |
Hb_004531_010 |
0.1447561325 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At2g01680-like [Jatropha curcas] |