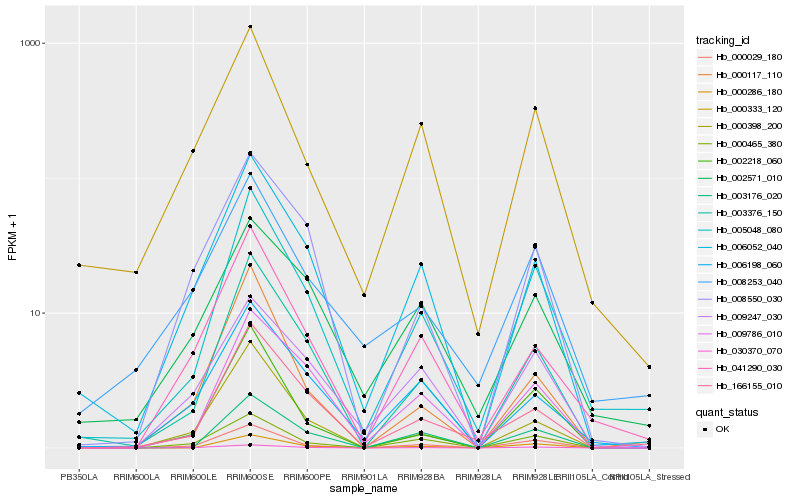
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005048_080 |
0.0 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 2 |
Hb_003376_150 |
0.1227540889 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 3 |
Hb_166155_010 |
0.1371350093 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 4 |
Hb_000029_180 |
0.1416992159 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 5 |
Hb_003176_020 |
0.1429217413 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 6 |
Hb_000286_180 |
0.1451906916 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 7 |
Hb_030370_070 |
0.1468557795 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_009786_010 |
0.1505928965 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 9 |
Hb_006052_040 |
0.1533882809 |
- |
- |
pyruvate decarboxylase [Hevea brasiliensis] |
| 10 |
Hb_041290_030 |
0.160730769 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 11 |
Hb_000465_380 |
0.1617640759 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_002218_060 |
0.1624053473 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000333_120 |
0.1671051061 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 14 |
Hb_008253_040 |
0.1727812473 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 15 |
Hb_000117_110 |
0.1734956367 |
- |
- |
transferase, putative [Ricinus communis] |
| 16 |
Hb_000398_200 |
0.1753085162 |
- |
- |
PREDICTED: uncharacterized protein LOC105631579 [Jatropha curcas] |
| 17 |
Hb_002571_010 |
0.1788388593 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_006198_060 |
0.1793837347 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 19 |
Hb_009247_030 |
0.181489837 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 20 |
Hb_008550_030 |
0.1835933995 |
- |
- |
PREDICTED: tonoplast dicarboxylate transporter [Jatropha curcas] |