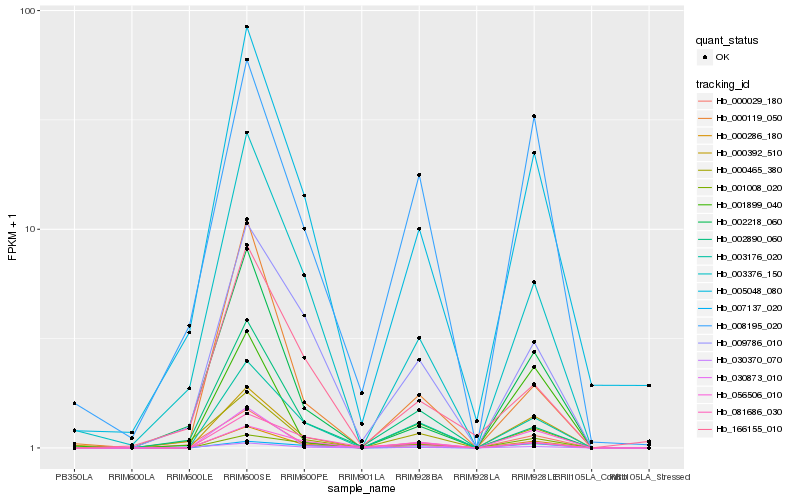
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000286_180 |
0.0 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 2 |
Hb_003176_020 |
0.1099913589 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 3 |
Hb_005048_080 |
0.1451906916 |
- |
- |
PREDICTED: PAP-specific phosphatase HAL2-like [Jatropha curcas] |
| 4 |
Hb_000392_510 |
0.1521424966 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 5 |
Hb_000029_180 |
0.153222794 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 6 |
Hb_003376_150 |
0.1548960435 |
- |
- |
PREDICTED: peroxidase 21 [Jatropha curcas] |
| 7 |
Hb_002218_060 |
0.1608944657 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_009786_010 |
0.1721326381 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 9 |
Hb_007137_020 |
0.1817446582 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 10 |
Hb_000119_050 |
0.1822546964 |
- |
- |
PREDICTED: transmembrane protein 45B-like [Jatropha curcas] |
| 11 |
Hb_000465_380 |
0.1847387166 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_030370_070 |
0.1852856541 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_056506_010 |
0.1896361996 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 14 |
Hb_030873_010 |
0.1917428126 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 15 isoform X2 [Cucumis sativus] |
| 15 |
Hb_008195_020 |
0.1959601631 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 16 |
Hb_081686_030 |
0.1966171825 |
- |
- |
hypothetical protein MIMGU_mgv11b016770mg [Erythranthe guttata] |
| 17 |
Hb_001008_020 |
0.1988985189 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 18 |
Hb_001899_040 |
0.2004732755 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 19 |
Hb_166155_010 |
0.2028992011 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 20 |
Hb_002890_060 |
0.2054956787 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |