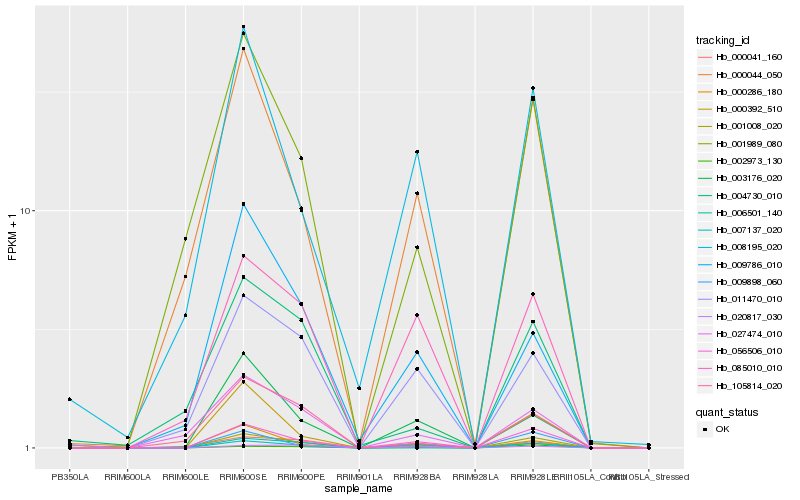
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007137_020 |
0.0 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 2 |
Hb_001008_020 |
0.0992981718 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 3 |
Hb_056506_010 |
0.1006798005 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 4 |
Hb_001989_080 |
0.1810111699 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 5 |
Hb_000286_180 |
0.1817446582 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 6 |
Hb_000392_510 |
0.1832655975 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 7 |
Hb_009898_060 |
0.1878508889 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 8 |
Hb_105814_020 |
0.1910394417 |
- |
- |
berberine bridge enzyme [Hevea brasiliensis] |
| 9 |
Hb_027474_010 |
0.1934050389 |
- |
- |
PREDICTED: uncharacterized protein LOC105629982 [Jatropha curcas] |
| 10 |
Hb_000044_050 |
0.1943106895 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000041_160 |
0.1971552527 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_006501_140 |
0.1994423576 |
- |
- |
PREDICTED: MATE efflux family protein 9-like [Jatropha curcas] |
| 13 |
Hb_003176_020 |
0.2004100505 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 14 |
Hb_002973_130 |
0.2021981905 |
- |
- |
PREDICTED: uncharacterized protein LOC104609908 [Nelumbo nucifera] |
| 15 |
Hb_004730_010 |
0.2036703674 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT5 [Jatropha curcas] |
| 16 |
Hb_011470_010 |
0.2040858733 |
- |
- |
hypothetical protein CISIN_1g022501mg [Citrus sinensis] |
| 17 |
Hb_085010_010 |
0.2043523236 |
- |
- |
PREDICTED: MLO-like protein 3 isoform X2 [Vitis vinifera] |
| 18 |
Hb_009786_010 |
0.2124775606 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 19 |
Hb_008195_020 |
0.2144897245 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 20 |
Hb_020817_030 |
0.2171461012 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |