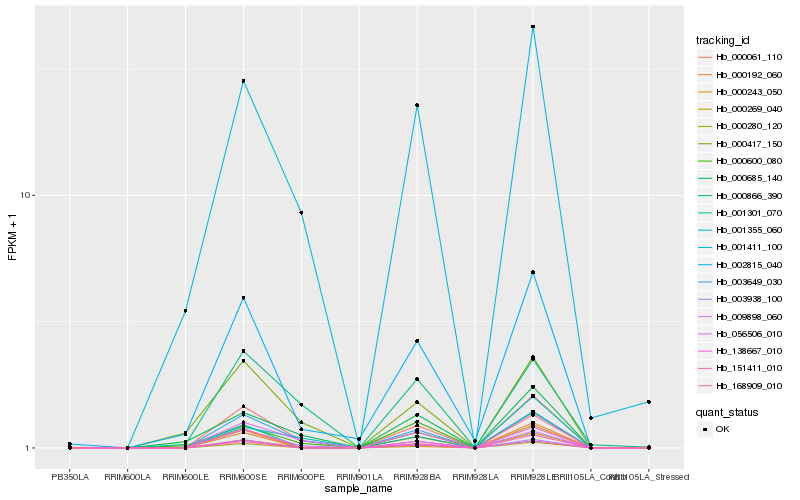
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000061_110 |
0.0 |
- |
- |
- |
| 2 |
Hb_003649_030 |
0.0563701642 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 3 |
Hb_009898_060 |
0.1339322635 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 4 |
Hb_002815_040 |
0.1498375001 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 5 |
Hb_001411_100 |
0.1505557019 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_001355_060 |
0.1522405522 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 7 |
Hb_056506_010 |
0.1603570444 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 8 |
Hb_000243_050 |
0.1615236574 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 9 |
Hb_003938_100 |
0.1640813779 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000417_150 |
0.1644282028 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 11 |
Hb_001301_070 |
0.1650516009 |
- |
- |
- |
| 12 |
Hb_000600_080 |
0.16988185 |
- |
- |
- |
| 13 |
Hb_000685_140 |
0.1701465269 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 14 |
Hb_000192_060 |
0.1720560453 |
- |
- |
- |
| 15 |
Hb_138667_010 |
0.1729289128 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 16 |
Hb_000280_120 |
0.1744704606 |
- |
- |
PREDICTED: uncharacterized protein LOC105643934 [Jatropha curcas] |
| 17 |
Hb_000269_040 |
0.1752253901 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_168909_010 |
0.1753482047 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 19 |
Hb_000866_390 |
0.1778136124 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 20 |
Hb_151411_010 |
0.1801882436 |
- |
- |
- |