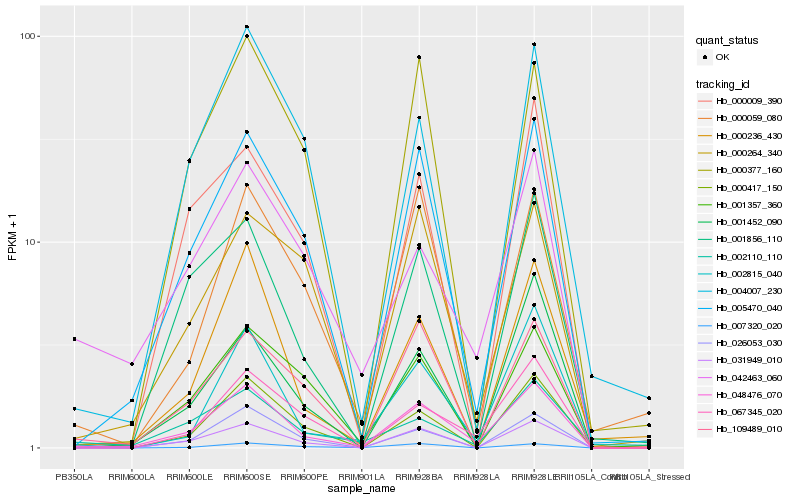
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_048476_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_005470_040 |
0.1170546866 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 3 |
Hb_031949_010 |
0.1370591603 |
- |
- |
- |
| 4 |
Hb_002815_040 |
0.1544192536 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 5 |
Hb_000417_150 |
0.154976513 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 6 |
Hb_026053_030 |
0.1577746634 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 7 |
Hb_067345_020 |
0.1589350128 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 8 |
Hb_000236_430 |
0.161081589 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 9 |
Hb_001856_110 |
0.1613742832 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 10 |
Hb_109489_010 |
0.1673368818 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 11 |
Hb_000059_080 |
0.1691578854 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 12 |
Hb_000377_160 |
0.1693057134 |
- |
- |
PREDICTED: oligopeptide transporter 3 [Jatropha curcas] |
| 13 |
Hb_001357_360 |
0.1763543347 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_004007_230 |
0.1773722638 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000264_340 |
0.1777040239 |
- |
- |
PREDICTED: callose synthase 11-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000009_390 |
0.1789142039 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001452_090 |
0.1796055221 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 18 |
Hb_007320_020 |
0.1799748541 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 isoform X2 [Populus euphratica] |
| 19 |
Hb_042463_060 |
0.1810513908 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002110_110 |
0.1812496969 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |