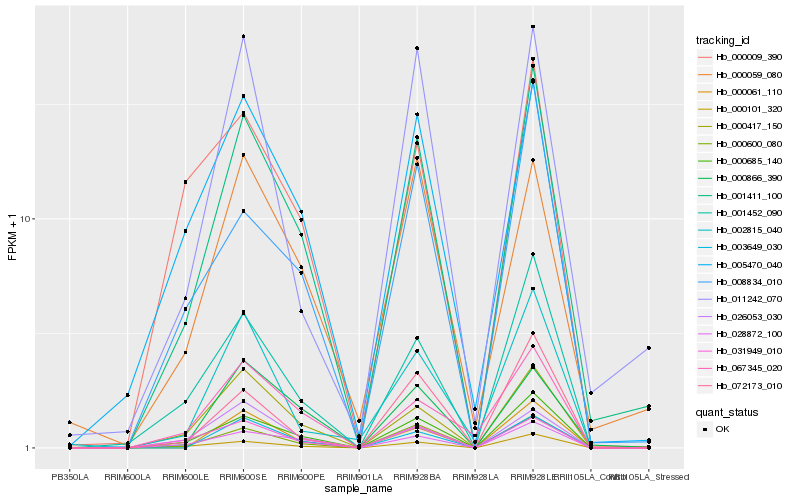
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001411_100 |
0.0 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 2 |
Hb_000685_140 |
0.0756275334 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 3 |
Hb_000600_080 |
0.1064225746 |
- |
- |
- |
| 4 |
Hb_000101_320 |
0.1085259755 |
- |
- |
- |
| 5 |
Hb_028872_100 |
0.1098910539 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 6 |
Hb_001452_090 |
0.1175913461 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 7 |
Hb_000417_150 |
0.1287605299 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 8 |
Hb_031949_010 |
0.1433741405 |
- |
- |
- |
| 9 |
Hb_000866_390 |
0.1449199656 |
- |
- |
PREDICTED: O-glucosyltransferase rumi homolog [Jatropha curcas] |
| 10 |
Hb_005470_040 |
0.146696498 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 11 |
Hb_000061_110 |
0.1505557019 |
- |
- |
- |
| 12 |
Hb_003649_030 |
0.1507593397 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 13 |
Hb_008834_010 |
0.1508355206 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |
| 14 |
Hb_072173_010 |
0.1545197966 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 15 |
Hb_000059_080 |
0.1568681343 |
- |
- |
PREDICTED: uncharacterized protein LOC105629177 [Jatropha curcas] |
| 16 |
Hb_011242_070 |
0.1611972959 |
- |
- |
beta-cyanoalanine synthase [Hevea brasiliensis] |
| 17 |
Hb_067345_020 |
0.1676290461 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 18 |
Hb_000009_390 |
0.1684652093 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_002815_040 |
0.1686407218 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 20 |
Hb_026053_030 |
0.1691929891 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |