| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
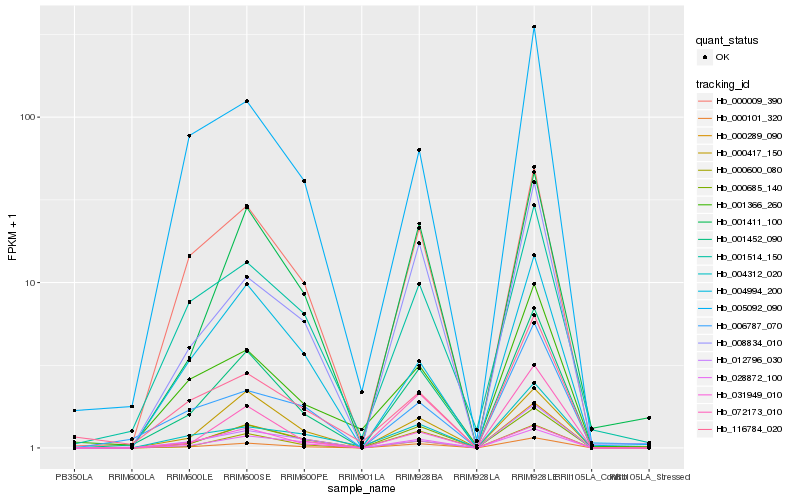
Hb_001452_090 |
0.0 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 2 |
Hb_000101_320 |
0.0509550466 |
- |
- |
- |
| 3 |
Hb_000685_140 |
0.0842105305 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 4 |
Hb_001411_100 |
0.1175913461 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 5 |
Hb_028872_100 |
0.1246223812 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 6 |
Hb_000009_390 |
0.1251563924 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 7 |
Hb_008834_010 |
0.13056873 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |
| 8 |
Hb_000417_150 |
0.1393574637 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 9 |
Hb_000600_080 |
0.1422264041 |
- |
- |
- |
| 10 |
Hb_031949_010 |
0.1444674085 |
- |
- |
- |
| 11 |
Hb_001514_150 |
0.1459522726 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 12 |
Hb_005092_090 |
0.1495481892 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 13 |
Hb_004312_020 |
0.1530384538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_072173_010 |
0.1534108951 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 15 |
Hb_000289_090 |
0.1548758863 |
- |
- |
- |
| 16 |
Hb_012796_030 |
0.1560375617 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 17 |
Hb_001366_260 |
0.156211427 |
- |
- |
T4.15 [Malus x robusta] |
| 18 |
Hb_116784_020 |
0.1567507004 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_004994_200 |
0.1574854084 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 20 |
Hb_006787_070 |
0.1584678673 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |