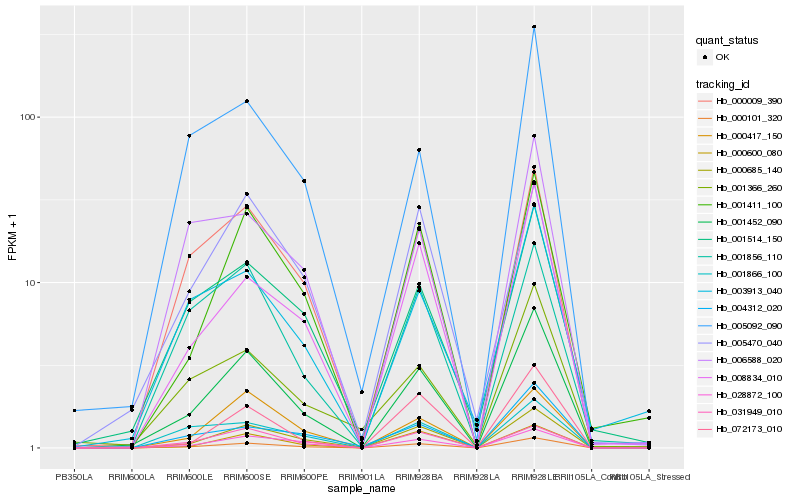
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000101_320 |
0.0 |
- |
- |
- |
| 2 |
Hb_001452_090 |
0.0509550466 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 3 |
Hb_000685_140 |
0.0609314975 |
- |
- |
PREDICTED: uncharacterized protein LOC105645669 [Jatropha curcas] |
| 4 |
Hb_008834_010 |
0.1067775399 |
- |
- |
hypothetical protein POPTR_0016s04660g [Populus trichocarpa] |
| 5 |
Hb_001411_100 |
0.1085259755 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 6 |
Hb_028872_100 |
0.1160669059 |
- |
- |
PREDICTED: uncharacterized protein LOC102707106 [Oryza brachyantha] |
| 7 |
Hb_000009_390 |
0.1160836171 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000600_080 |
0.1210060375 |
- |
- |
- |
| 9 |
Hb_031949_010 |
0.1295318158 |
- |
- |
- |
| 10 |
Hb_072173_010 |
0.1362956195 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 34 [Jatropha curcas] |
| 11 |
Hb_004312_020 |
0.1410441122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000417_150 |
0.1454382277 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 13 |
Hb_001866_100 |
0.1476059138 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 14 |
Hb_001514_150 |
0.1506908441 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 15 |
Hb_001856_110 |
0.1552320001 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |
| 16 |
Hb_006588_020 |
0.1566790916 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_005092_090 |
0.1580501592 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 18 |
Hb_005470_040 |
0.158620891 |
- |
- |
hypothetical protein POPTR_0015s09760g [Populus trichocarpa] |
| 19 |
Hb_001366_260 |
0.1610784968 |
- |
- |
T4.15 [Malus x robusta] |
| 20 |
Hb_003913_040 |
0.1628237625 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |