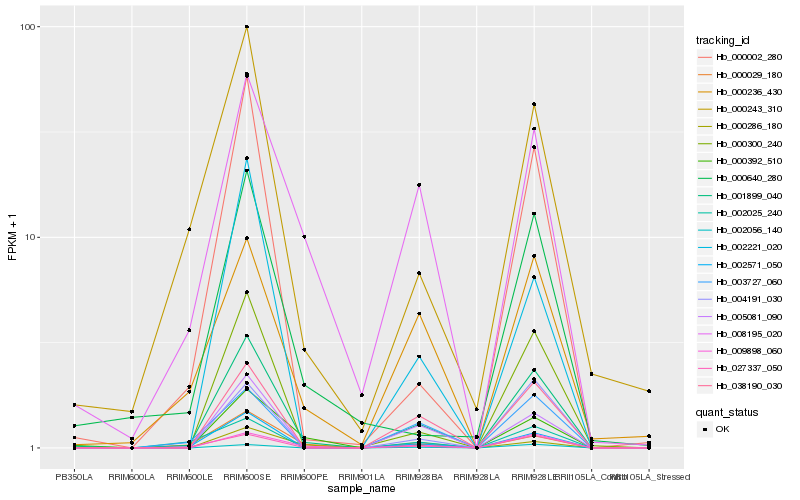
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001899_040 |
0.0 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 2 |
Hb_002571_050 |
0.1325006818 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 3 |
Hb_027337_050 |
0.1336677963 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_009898_060 |
0.14706374 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 5 |
Hb_000300_240 |
0.1481357342 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 6 |
Hb_038190_030 |
0.1528059037 |
- |
- |
- |
| 7 |
Hb_000002_280 |
0.1616328618 |
- |
- |
PREDICTED: two-pore potassium channel 1 [Jatropha curcas] |
| 8 |
Hb_005081_090 |
0.1663243032 |
- |
- |
- |
| 9 |
Hb_002056_140 |
0.1774533712 |
- |
- |
- |
| 10 |
Hb_004191_030 |
0.1822621419 |
- |
- |
hypothetical protein POPTR_0013s13140g [Populus trichocarpa] |
| 11 |
Hb_000392_510 |
0.1846961515 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Jatropha curcas] |
| 12 |
Hb_003727_060 |
0.1847690347 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 13 |
Hb_000243_310 |
0.18870084 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 14 |
Hb_008195_020 |
0.1900047893 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 15 |
Hb_000029_180 |
0.1904211775 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_002221_020 |
0.1917059871 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 17 |
Hb_000286_180 |
0.2004732755 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 4 [Jatropha curcas] |
| 18 |
Hb_000236_430 |
0.2007960042 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 19 |
Hb_002025_240 |
0.2051975405 |
- |
- |
- |
| 20 |
Hb_000640_280 |
0.206081391 |
- |
- |
Stachyose synthase precursor, putative [Ricinus communis] |