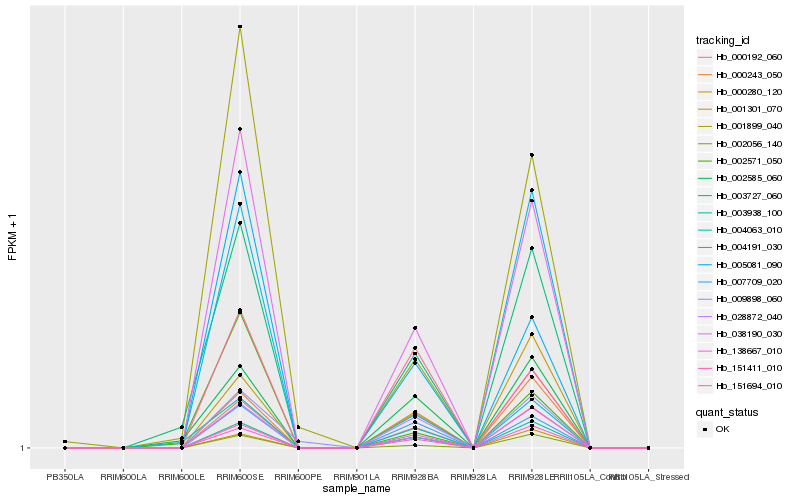
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_038190_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_003938_100 |
0.0888240691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005081_090 |
0.1121805118 |
- |
- |
- |
| 4 |
Hb_002056_140 |
0.1122303171 |
- |
- |
- |
| 5 |
Hb_007709_020 |
0.1216371876 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 6 |
Hb_003727_060 |
0.130989935 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_138667_010 |
0.1393359338 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 8 |
Hb_028872_040 |
0.1419410631 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_000243_050 |
0.1431715826 |
- |
- |
ADP-ribosylation factor, putative [Ricinus communis] |
| 10 |
Hb_001899_040 |
0.1528059037 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 11 |
Hb_004063_010 |
0.1619477496 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_004191_030 |
0.1671000268 |
- |
- |
hypothetical protein POPTR_0013s13140g [Populus trichocarpa] |
| 13 |
Hb_000280_120 |
0.1698598342 |
- |
- |
PREDICTED: uncharacterized protein LOC105643934 [Jatropha curcas] |
| 14 |
Hb_001301_070 |
0.170423874 |
- |
- |
- |
| 15 |
Hb_002585_060 |
0.1749225306 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_000192_060 |
0.1777195624 |
- |
- |
- |
| 17 |
Hb_009898_060 |
0.1804403389 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 18 |
Hb_002571_050 |
0.1828356274 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 19 |
Hb_151694_010 |
0.1829836247 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_151411_010 |
0.1863718443 |
- |
- |
- |