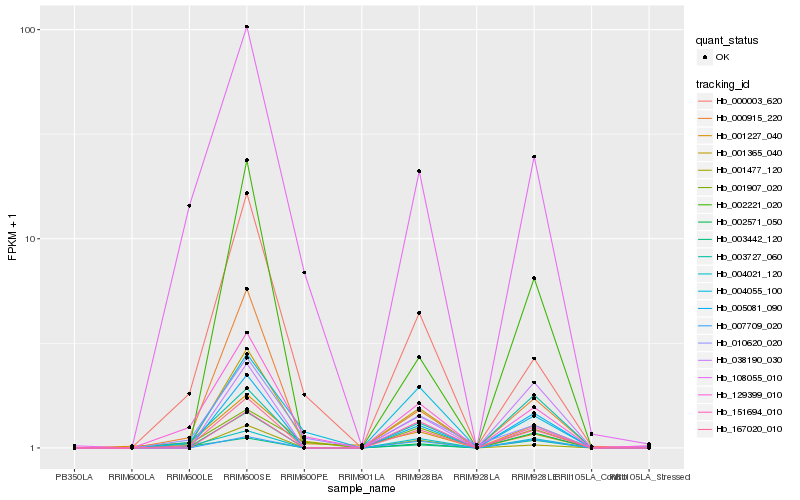
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167020_010 |
0.0 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 2 |
Hb_005081_090 |
0.1175550329 |
- |
- |
- |
| 3 |
Hb_004021_120 |
0.1357674023 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 4 |
Hb_000915_220 |
0.1600537572 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 5 |
Hb_001477_120 |
0.1619595718 |
- |
- |
hypothetical protein JCGZ_25721 [Jatropha curcas] |
| 6 |
Hb_002571_050 |
0.1629319058 |
- |
- |
PREDICTED: cation/calcium exchanger 1 [Jatropha curcas] |
| 7 |
Hb_129399_010 |
0.1758448966 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 8 |
Hb_151694_010 |
0.1805579235 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_038190_030 |
0.1870313283 |
- |
- |
- |
| 10 |
Hb_003727_060 |
0.1934218017 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 11 |
Hb_007709_020 |
0.1944226416 |
- |
- |
PREDICTED: uncharacterized protein LOC105049010 [Elaeis guineensis] |
| 12 |
Hb_108055_010 |
0.1998923252 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 13 |
Hb_002221_020 |
0.2001484315 |
- |
- |
hypothetical protein B456_008G083700 [Gossypium raimondii] |
| 14 |
Hb_000003_620 |
0.2003116299 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 15 |
Hb_001227_040 |
0.2015447746 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 16 |
Hb_001365_040 |
0.202045133 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 17 |
Hb_001907_020 |
0.2022172976 |
- |
- |
PREDICTED: receptor-like protein 12 [Vitis vinifera] |
| 18 |
Hb_010620_020 |
0.2031684767 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 19 |
Hb_004055_100 |
0.2045273413 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Jatropha curcas] |
| 20 |
Hb_003442_120 |
0.205285485 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |