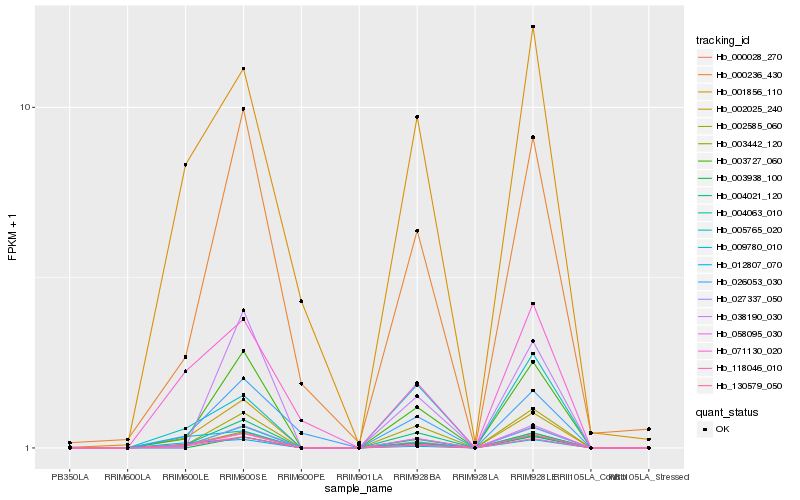
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_118046_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 2 |
Hb_003442_120 |
0.0253635481 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 3 |
Hb_004063_010 |
0.0612547812 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_003727_060 |
0.08430272 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_130579_050 |
0.0868375002 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 6 |
Hb_002025_240 |
0.0906856549 |
- |
- |
- |
| 7 |
Hb_000028_270 |
0.0971521005 |
- |
- |
- |
| 8 |
Hb_002585_060 |
0.1260133026 |
- |
- |
PREDICTED: uncharacterized protein LOC104899128, partial [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_058095_030 |
0.1336564603 |
- |
- |
- |
| 10 |
Hb_004021_120 |
0.1464633067 |
- |
- |
PREDICTED: secoisolariciresinol dehydrogenase-like [Jatropha curcas] |
| 11 |
Hb_000236_430 |
0.1604239181 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 12 |
Hb_027337_050 |
0.1810514593 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_012807_070 |
0.1941600715 |
- |
- |
PREDICTED: UDP-glycosyltransferase 86A2 [Jatropha curcas] |
| 14 |
Hb_026053_030 |
0.1946284777 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 15 |
Hb_003938_100 |
0.198184792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_071130_020 |
0.1987566271 |
- |
- |
- |
| 17 |
Hb_038190_030 |
0.1988016283 |
- |
- |
- |
| 18 |
Hb_009780_010 |
0.2002186526 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005765_020 |
0.2055733149 |
- |
- |
- |
| 20 |
Hb_001856_110 |
0.206333691 |
- |
- |
PREDICTED: cellulose synthase-like protein G2 [Populus euphratica] |