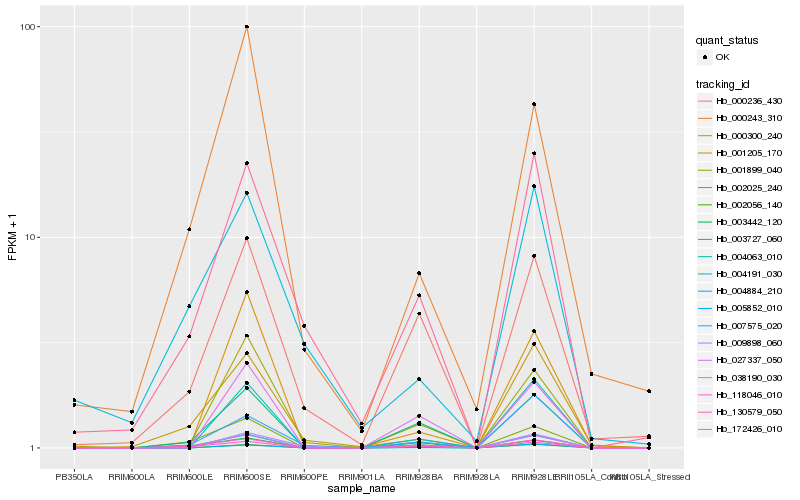
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027337_050 |
0.0 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 2 |
Hb_001899_040 |
0.1336677963 |
- |
- |
PREDICTED: cyclic nucleotide-gated ion channel 2-like [Jatropha curcas] |
| 3 |
Hb_130579_050 |
0.1500752568 |
- |
- |
hypothetical protein JCGZ_25577 [Jatropha curcas] |
| 4 |
Hb_172426_010 |
0.1536659578 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 5 |
Hb_002025_240 |
0.1627962681 |
- |
- |
- |
| 6 |
Hb_001205_170 |
0.1637242712 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003727_060 |
0.1674133012 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_007575_020 |
0.1714835474 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_004191_030 |
0.1730493723 |
- |
- |
hypothetical protein POPTR_0013s13140g [Populus trichocarpa] |
| 10 |
Hb_004063_010 |
0.1731097983 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 11 |
Hb_002056_140 |
0.1761067243 |
- |
- |
- |
| 12 |
Hb_003442_120 |
0.1800469129 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 13 |
Hb_118046_010 |
0.1810514593 |
- |
- |
PREDICTED: uncharacterized protein LOC102596706 [Solanum tuberosum] |
| 14 |
Hb_009898_060 |
0.1849976721 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 15 |
Hb_005852_010 |
0.1868990799 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_000236_430 |
0.1902811448 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 17 |
Hb_000243_310 |
0.1991378274 |
- |
- |
lactoylglutathione lyase, putative [Ricinus communis] |
| 18 |
Hb_004884_210 |
0.2012843551 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 19 |
Hb_000300_240 |
0.2049216768 |
- |
- |
Sodium/potassium/calcium exchanger 6 precursor, putative [Ricinus communis] |
| 20 |
Hb_038190_030 |
0.2061754353 |
- |
- |
- |