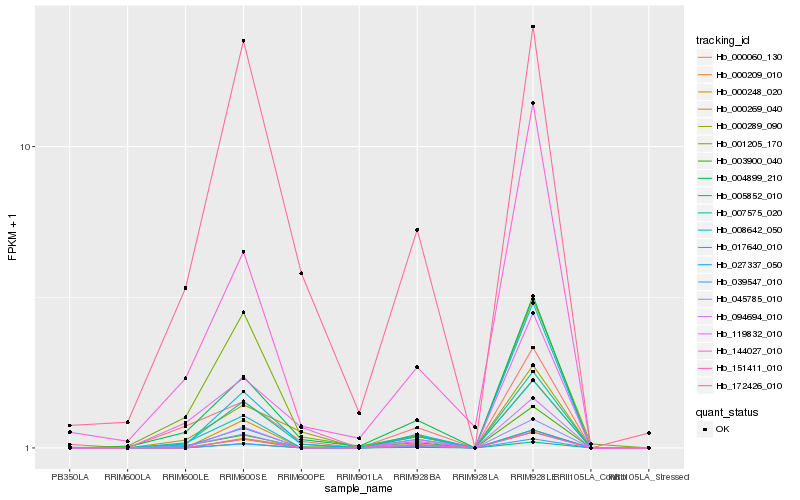
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007575_020 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_000269_040 |
0.1106139519 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 3 |
Hb_004899_210 |
0.1173461306 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 4 |
Hb_045785_010 |
0.1189565233 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 5 |
Hb_000289_090 |
0.139671954 |
- |
- |
- |
| 6 |
Hb_008642_050 |
0.1409615887 |
- |
- |
- |
| 7 |
Hb_017640_010 |
0.1464138852 |
- |
- |
hypothetical protein JCGZ_18815 [Jatropha curcas] |
| 8 |
Hb_003900_040 |
0.1556149588 |
- |
- |
hypothetical protein [Lotus japonicus] |
| 9 |
Hb_001205_170 |
0.1585201759 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 10 |
Hb_119832_010 |
0.1604312356 |
- |
- |
- |
| 11 |
Hb_005852_010 |
0.162697868 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_172426_010 |
0.1645502798 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 13 |
Hb_027337_050 |
0.1714835474 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 14 |
Hb_144027_010 |
0.1726839269 |
- |
- |
- |
| 15 |
Hb_094694_010 |
0.1782340651 |
- |
- |
- |
| 16 |
Hb_151411_010 |
0.178739039 |
- |
- |
- |
| 17 |
Hb_000248_020 |
0.1808099014 |
- |
- |
PREDICTED: uncharacterized protein LOC104243878 [Nicotiana sylvestris] |
| 18 |
Hb_000060_130 |
0.1808367352 |
- |
- |
- |
| 19 |
Hb_000209_010 |
0.1811433366 |
- |
- |
- |
| 20 |
Hb_039547_010 |
0.1832804781 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |