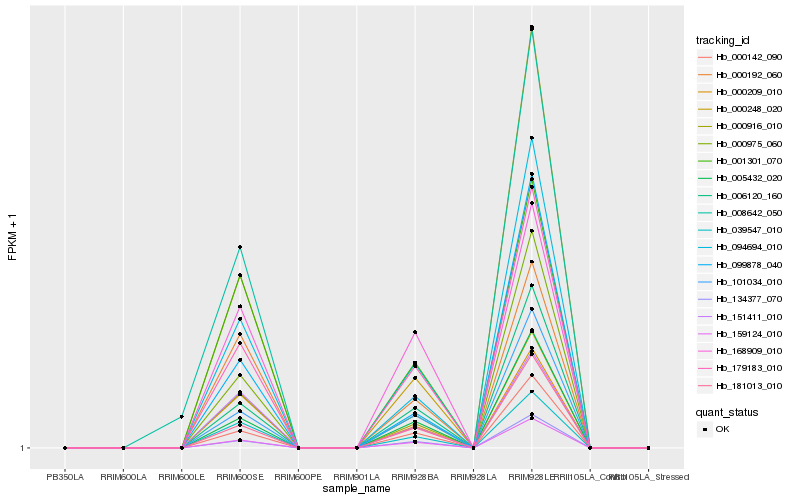
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039547_010 |
0.0 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 2 |
Hb_000916_010 |
0.0009126977 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 3 |
Hb_000209_010 |
0.0274344543 |
- |
- |
- |
| 4 |
Hb_151411_010 |
0.0436708698 |
- |
- |
- |
| 5 |
Hb_094694_010 |
0.0489826508 |
- |
- |
- |
| 6 |
Hb_000192_060 |
0.0519366808 |
- |
- |
- |
| 7 |
Hb_000248_020 |
0.0611949899 |
- |
- |
PREDICTED: uncharacterized protein LOC104243878 [Nicotiana sylvestris] |
| 8 |
Hb_001301_070 |
0.0681642034 |
- |
- |
- |
| 9 |
Hb_179183_010 |
0.0725797013 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 10 |
Hb_000975_060 |
0.0764788325 |
- |
- |
- |
| 11 |
Hb_159124_010 |
0.0938641212 |
- |
- |
PREDICTED: uncharacterized protein LOC103332224 [Prunus mume] |
| 12 |
Hb_099878_040 |
0.0983424416 |
- |
- |
PREDICTED: uncharacterized protein LOC102703670 [Oryza brachyantha] |
| 13 |
Hb_006120_160 |
0.1061506219 |
- |
- |
- |
| 14 |
Hb_101034_010 |
0.1088058345 |
- |
- |
- |
| 15 |
Hb_005432_020 |
0.1113991775 |
- |
- |
- |
| 16 |
Hb_181013_010 |
0.1144600297 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |
| 17 |
Hb_000142_090 |
0.1171469031 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 18 |
Hb_168909_010 |
0.1205726243 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 19 |
Hb_134377_070 |
0.1222484363 |
- |
- |
- |
| 20 |
Hb_008642_050 |
0.136667809 |
- |
- |
- |