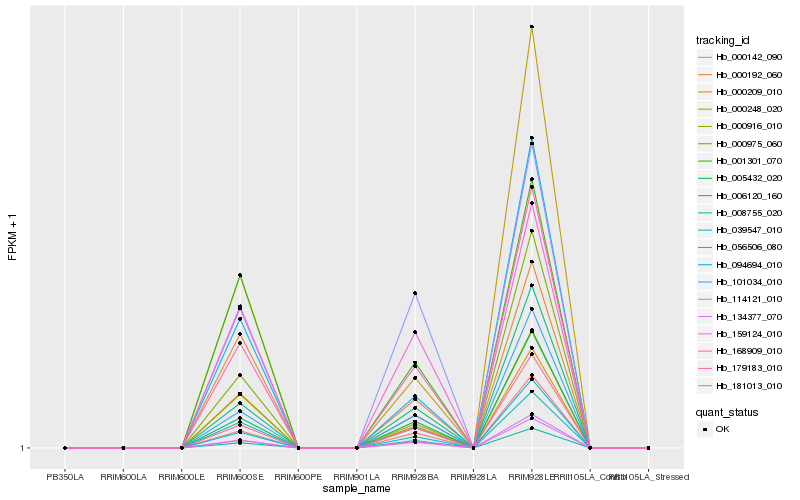
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179183_010 |
0.0 |
- |
- |
RALF-LIKE 32 family protein [Populus trichocarpa] |
| 2 |
Hb_006120_160 |
0.060795796 |
- |
- |
- |
| 3 |
Hb_101034_010 |
0.0670942832 |
- |
- |
- |
| 4 |
Hb_159124_010 |
0.072106636 |
- |
- |
PREDICTED: uncharacterized protein LOC103332224 [Prunus mume] |
| 5 |
Hb_000916_010 |
0.072521958 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 6 |
Hb_039547_010 |
0.0725797013 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 3-like [Jatropha curcas] |
| 7 |
Hb_005432_020 |
0.0725896562 |
- |
- |
- |
| 8 |
Hb_181013_010 |
0.0785176324 |
- |
- |
PREDICTED: uncharacterized protein LOC105629028 [Jatropha curcas] |
| 9 |
Hb_008755_020 |
0.0822314269 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 10 |
Hb_114121_010 |
0.0828258497 |
- |
- |
- |
| 11 |
Hb_056506_080 |
0.0829246474 |
- |
- |
PREDICTED: uncharacterized protein LOC104888944 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_000142_090 |
0.0833593137 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 [Jatropha curcas] |
| 13 |
Hb_168909_010 |
0.0881264179 |
- |
- |
PREDICTED: uncharacterized protein LOC105640156 [Jatropha curcas] |
| 14 |
Hb_000209_010 |
0.0882591822 |
- |
- |
- |
| 15 |
Hb_134377_070 |
0.0918906076 |
- |
- |
- |
| 16 |
Hb_001301_070 |
0.0947483215 |
- |
- |
- |
| 17 |
Hb_094694_010 |
0.0962483843 |
- |
- |
- |
| 18 |
Hb_000192_060 |
0.0973915544 |
- |
- |
- |
| 19 |
Hb_000975_060 |
0.1014619884 |
- |
- |
- |
| 20 |
Hb_000248_020 |
0.1024677374 |
- |
- |
PREDICTED: uncharacterized protein LOC104243878 [Nicotiana sylvestris] |