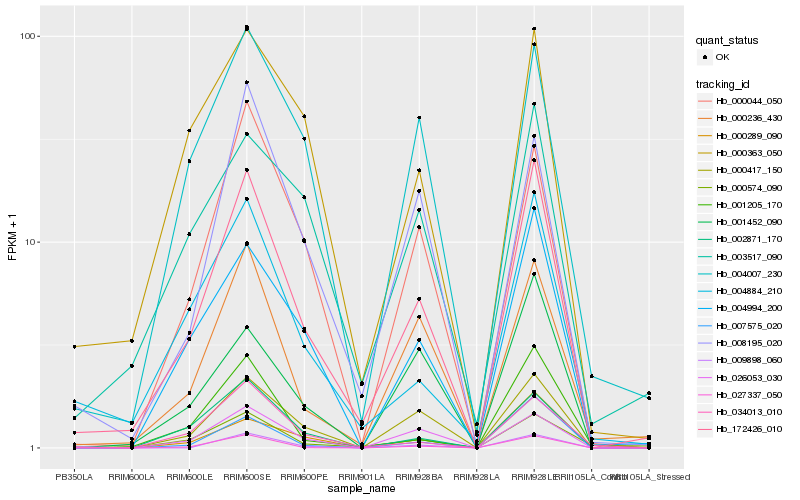
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172426_010 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 2 |
Hb_000417_150 |
0.1359186084 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 3 |
Hb_004994_200 |
0.1377326908 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 4 |
Hb_000236_430 |
0.1485121308 |
- |
- |
PREDICTED: uncharacterized protein LOC105633302 [Jatropha curcas] |
| 5 |
Hb_004884_210 |
0.1487179806 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 6 |
Hb_027337_050 |
0.1536659578 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 7 |
Hb_001205_170 |
0.1585836939 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000574_090 |
0.1607904415 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 9 |
Hb_026053_030 |
0.1611941897 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |
| 10 |
Hb_000044_050 |
0.1624785795 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_007575_020 |
0.1645502798 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 12 |
Hb_002871_170 |
0.1662206712 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 13 |
Hb_000289_090 |
0.1670483236 |
- |
- |
- |
| 14 |
Hb_004007_230 |
0.1688180356 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000363_050 |
0.1715759575 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_034013_010 |
0.1766256846 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 17 |
Hb_009898_060 |
0.1771310134 |
- |
- |
hypothetical protein POPTR_0007s09865g [Populus trichocarpa] |
| 18 |
Hb_008195_020 |
0.1773389592 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 19 |
Hb_001452_090 |
0.1800444272 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 20 |
Hb_003517_090 |
0.1824483723 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |