| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
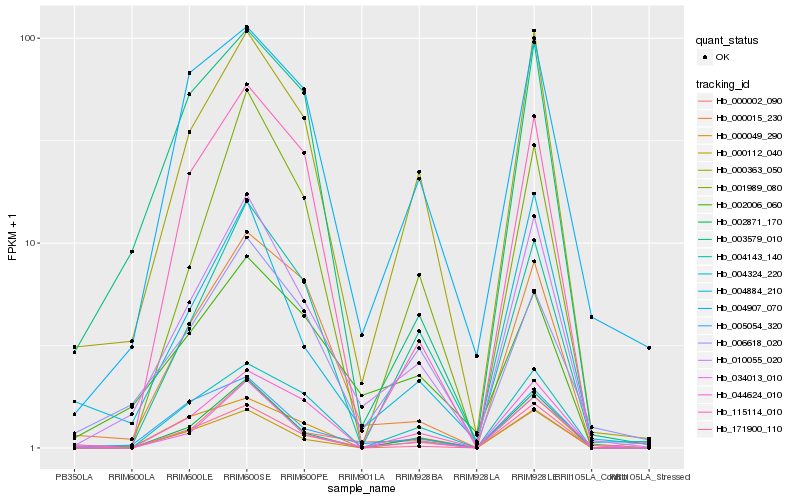
Hb_010055_020 |
0.0 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 2 |
Hb_000363_050 |
0.1073429947 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_000002_090 |
0.1344692387 |
- |
- |
PREDICTED: RING-H2 finger protein ATL8-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002871_170 |
0.1369983643 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |
| 5 |
Hb_005054_320 |
0.1375470982 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 6 |
Hb_034013_010 |
0.1417479731 |
- |
- |
hypothetical protein JCGZ_21944 [Jatropha curcas] |
| 7 |
Hb_004884_210 |
0.1456120042 |
- |
- |
PREDICTED: mitoferrin-like [Jatropha curcas] |
| 8 |
Hb_004143_140 |
0.146562993 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 9 |
Hb_000112_040 |
0.1504943496 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_000015_230 |
0.1560859651 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 11 |
Hb_002006_060 |
0.1561546071 |
- |
- |
PREDICTED: oleosin 1-like [Jatropha curcas] |
| 12 |
Hb_004907_070 |
0.1564644746 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 13 |
Hb_044624_010 |
0.1565328379 |
- |
- |
- |
| 14 |
Hb_000049_290 |
0.1577092007 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 15 |
Hb_001989_080 |
0.1583889692 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_006618_020 |
0.1634219225 |
- |
- |
Vacuolar-processing enzyme precursor [Ricinus communis] |
| 17 |
Hb_171900_110 |
0.1641969473 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 18 |
Hb_004324_220 |
0.1648474225 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 19 |
Hb_115114_010 |
0.1664920198 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003579_010 |
0.1675179052 |
- |
- |
cryptochrome 1.2 [Populus tremula] |