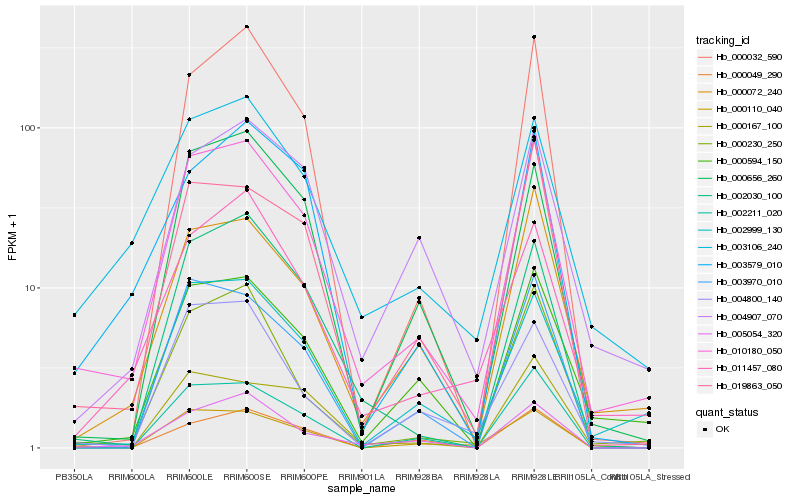
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010180_050 |
0.0 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 2 |
Hb_000072_240 |
0.114253723 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000594_150 |
0.1176770844 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN7, chloroplastic-like isoform X1 [Nelumbo nucifera] |
| 4 |
Hb_002999_130 |
0.1183887578 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 5 |
Hb_005054_320 |
0.1217642025 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 6 |
Hb_000230_250 |
0.1257080991 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003106_240 |
0.1297871113 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 8 |
Hb_002030_100 |
0.1312976157 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 9 |
Hb_000656_260 |
0.1315002765 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 10 |
Hb_011457_080 |
0.1339406759 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002211_020 |
0.1345411869 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 12 |
Hb_019863_050 |
0.1375140284 |
- |
- |
PREDICTED: uncharacterized protein LOC103343426 [Prunus mume] |
| 13 |
Hb_000032_590 |
0.1380847182 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 14 |
Hb_004907_070 |
0.1433184835 |
- |
- |
PREDICTED: protein phosphatase 2C 37 [Jatropha curcas] |
| 15 |
Hb_003970_010 |
0.1433535905 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_003579_010 |
0.1444593661 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 17 |
Hb_000110_040 |
0.1453918439 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 18 |
Hb_000049_290 |
0.1456935443 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 19 |
Hb_000167_100 |
0.1498916934 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 20 |
Hb_004800_140 |
0.1504543573 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |