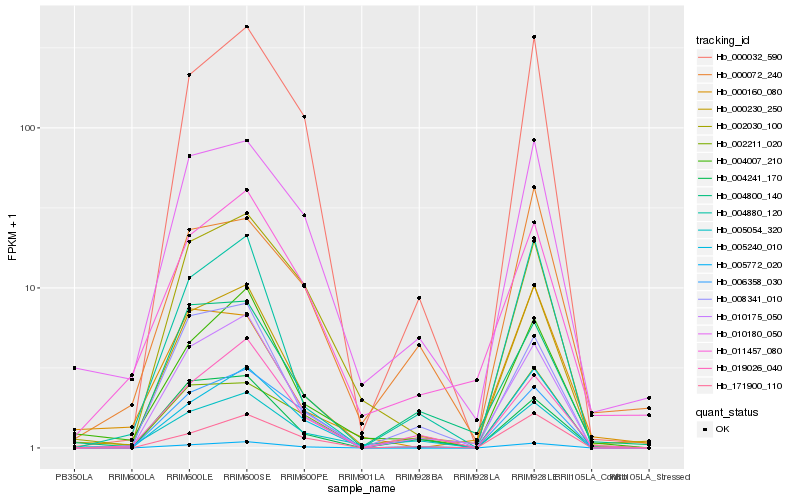
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_250 |
0.0 |
- |
- |
PREDICTED: pyruvate, phosphate dikinase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004880_120 |
0.11692421 |
- |
- |
PREDICTED: calcium-binding protein PBP1 [Jatropha curcas] |
| 3 |
Hb_005772_020 |
0.123397187 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 4 |
Hb_010175_050 |
0.1238244548 |
- |
- |
PREDICTED: uncharacterized protein LOC105639906 [Jatropha curcas] |
| 5 |
Hb_010180_050 |
0.1257080991 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 6 |
Hb_004007_210 |
0.1310332009 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000032_590 |
0.1341702538 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 8 |
Hb_005240_010 |
0.1457498521 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 9 |
Hb_000160_080 |
0.1529575178 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM2b-like [Jatropha curcas] |
| 10 |
Hb_004800_140 |
0.153479201 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 11 |
Hb_171900_110 |
0.1549468194 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 12 |
Hb_005054_320 |
0.156258474 |
- |
- |
PREDICTED: uncharacterized protein LOC105646518 [Jatropha curcas] |
| 13 |
Hb_000072_240 |
0.1583937231 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_008341_010 |
0.1617320521 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_002211_020 |
0.1649002796 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 16 |
Hb_011457_080 |
0.1668449621 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_019026_040 |
0.1726290194 |
- |
- |
S-locus lectin protein kinase family protein [Theobroma cacao] |
| 18 |
Hb_006358_030 |
0.1733718082 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 19 |
Hb_002030_100 |
0.1756631488 |
- |
- |
PREDICTED: putative clathrin assembly protein At4g40080 [Jatropha curcas] |
| 20 |
Hb_004241_170 |
0.1757795064 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |