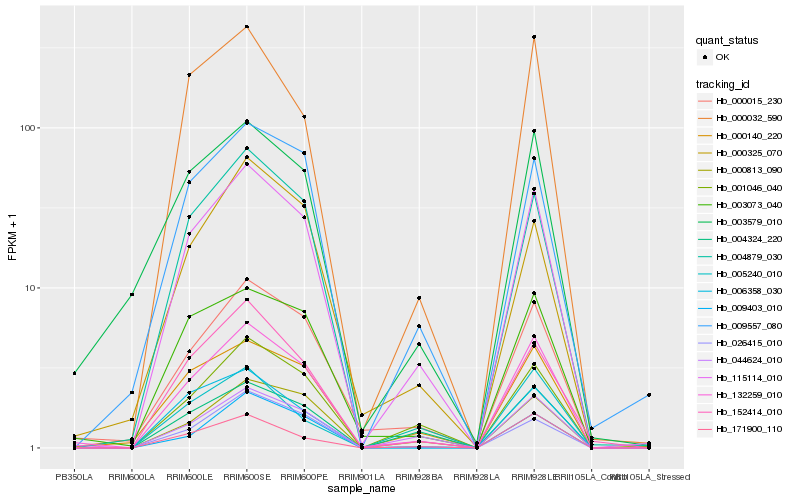
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132259_010 |
0.0 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 2 |
Hb_115114_010 |
0.06841814 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000015_230 |
0.0884489669 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 4 |
Hb_003073_040 |
0.10748746 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 5 |
Hb_004879_030 |
0.1131032529 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 6 |
Hb_000140_220 |
0.1139663171 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 7 |
Hb_000032_590 |
0.1201968232 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 8 |
Hb_000813_090 |
0.1249900078 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 9 |
Hb_003579_010 |
0.1262667419 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 10 |
Hb_044624_010 |
0.126988119 |
- |
- |
- |
| 11 |
Hb_152414_010 |
0.1276764823 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein PRUPE_ppa024232mg [Prunus persica] |
| 12 |
Hb_171900_110 |
0.1284133406 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT3-like [Populus euphratica] |
| 13 |
Hb_006358_030 |
0.1318681183 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 14 |
Hb_001046_040 |
0.1326430705 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 15 |
Hb_026415_010 |
0.1368893573 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 16 |
Hb_009557_080 |
0.1386043792 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 17 |
Hb_004324_220 |
0.1410874501 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 18 |
Hb_000325_070 |
0.141615851 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_009403_010 |
0.1432139829 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 20 |
Hb_005240_010 |
0.148320608 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |