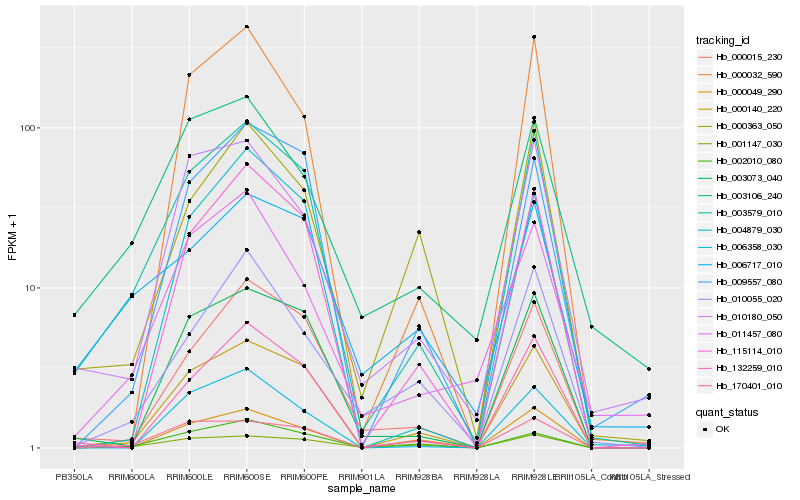
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003579_010 |
0.0 |
- |
- |
cryptochrome 1.2 [Populus tremula] |
| 2 |
Hb_000140_220 |
0.090660804 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 3 |
Hb_003073_040 |
0.1262174627 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 4 |
Hb_132259_010 |
0.1262667419 |
- |
- |
PREDICTED: WAT1-related protein At5g64700 [Jatropha curcas] |
| 5 |
Hb_000015_230 |
0.1304427706 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 6 |
Hb_115114_010 |
0.1348018423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000049_290 |
0.1432028548 |
- |
- |
PREDICTED: multiple C2 and transmembrane domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_009557_080 |
0.1436676637 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 9 |
Hb_010180_050 |
0.1444593661 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 10 |
Hb_000032_590 |
0.1512623706 |
- |
- |
PREDICTED: glutaredoxin domain-containing cysteine-rich protein 1 [Jatropha curcas] |
| 11 |
Hb_002010_080 |
0.1566631277 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g11050 [Jatropha curcas] |
| 12 |
Hb_006358_030 |
0.1572259842 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 13 |
Hb_004879_030 |
0.157777619 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 14 |
Hb_006717_010 |
0.1605452142 |
- |
- |
PREDICTED: uncharacterized protein LOC105649338 [Jatropha curcas] |
| 15 |
Hb_001147_030 |
0.1626709214 |
- |
- |
hypothetical protein JCGZ_14809 [Jatropha curcas] |
| 16 |
Hb_011457_080 |
0.1636953484 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003106_240 |
0.166085963 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 18 |
Hb_170401_010 |
0.1668107483 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 19 |
Hb_010055_020 |
0.1675179052 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 20 |
Hb_000363_050 |
0.16773073 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |