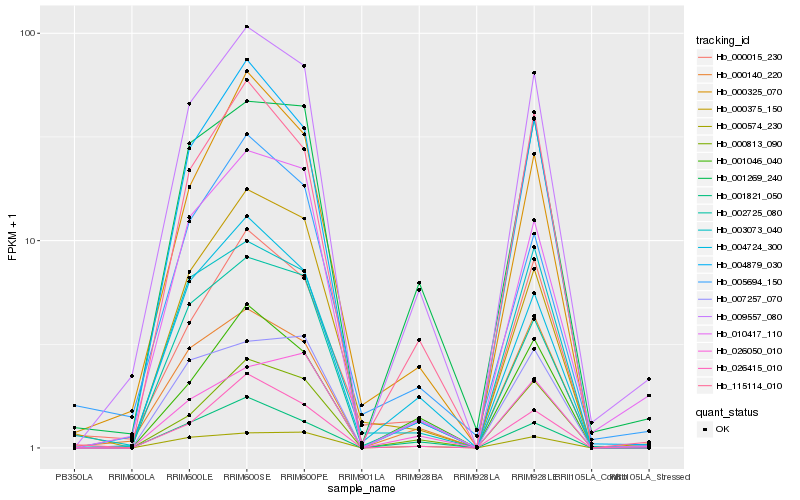
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009557_080 |
0.0 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 2 |
Hb_002725_080 |
0.0965867516 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 3 |
Hb_001269_240 |
0.0980634125 |
- |
- |
aquaporin NIP6.1 family protein [Populus trichocarpa] |
| 4 |
Hb_115114_010 |
0.0985879569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010417_110 |
0.0991167946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004879_030 |
0.101466391 |
- |
- |
PREDICTED: uncharacterized protein LOC105124685 [Populus euphratica] |
| 7 |
Hb_000140_220 |
0.1017526174 |
transcription factor |
TF Family: WRKY |
PREDICTED: WRKY transcription factor 22 [Jatropha curcas] |
| 8 |
Hb_000813_090 |
0.1021818464 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 9 |
Hb_004724_300 |
0.106736428 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 90-like [Jatropha curcas] |
| 10 |
Hb_001046_040 |
0.1096911729 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 11 |
Hb_000325_070 |
0.1101206134 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_001821_050 |
0.114910154 |
- |
- |
hypothetical protein CICLE_v10032084mg [Citrus clementina] |
| 13 |
Hb_026415_010 |
0.1173574397 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000574_230 |
0.117961291 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EXS-like isoform X2 [Vitis vinifera] |
| 15 |
Hb_000015_230 |
0.1252434352 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 16 |
Hb_007257_070 |
0.1287116911 |
- |
- |
PREDICTED: uncharacterized protein LOC105638098 [Jatropha curcas] |
| 17 |
Hb_026050_010 |
0.1316890097 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 18 |
Hb_003073_040 |
0.1330232986 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: dof zinc finger protein DOF3.6-like isoform X2 [Populus euphratica] |
| 19 |
Hb_005694_150 |
0.1331030779 |
- |
- |
PREDICTED: probable mitochondrial chaperone BCS1-A [Jatropha curcas] |
| 20 |
Hb_000375_150 |
0.1342744624 |
- |
- |
conserved hypothetical protein [Ricinus communis] |