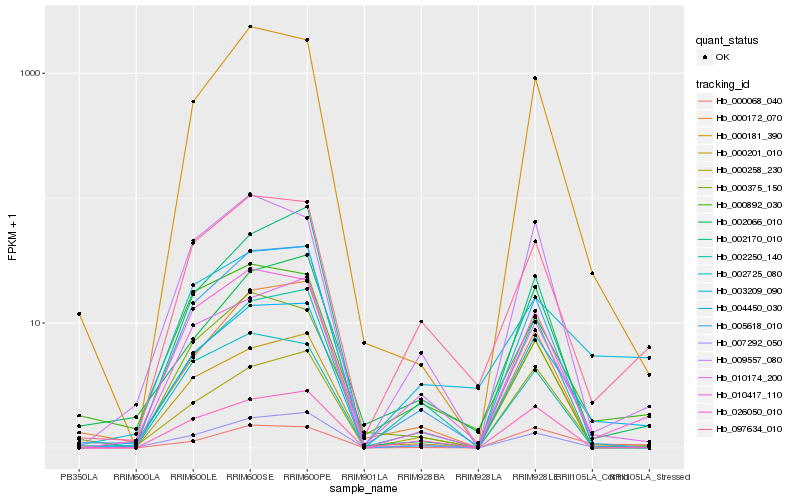
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004450_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_010417_110 |
0.0999235395 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000181_390 |
0.1210550089 |
- |
- |
unknown [Lotus japonicus] |
| 4 |
Hb_005618_010 |
0.1362851079 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0014s10680g [Populus trichocarpa] |
| 5 |
Hb_000258_230 |
0.1469187611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000068_040 |
0.1496834041 |
- |
- |
Inter-alpha-trypsin inhibitor heavy chain H3 precursor, putative [Ricinus communis] |
| 7 |
Hb_097634_010 |
0.1502649491 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 8 |
Hb_007292_050 |
0.1564337382 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance RPP13-like protein 4 [Jatropha curcas] |
| 9 |
Hb_010174_200 |
0.1565833792 |
- |
- |
PREDICTED: uncharacterized protein LOC105644743 isoform X1 [Jatropha curcas] |
| 10 |
Hb_009557_080 |
0.1570638381 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 11 |
Hb_026050_010 |
0.1578889151 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 12 |
Hb_002725_080 |
0.1625196234 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 13 |
Hb_000375_150 |
0.1627115623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000172_070 |
0.1654318349 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 15 |
Hb_003209_090 |
0.1668246608 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 16 |
Hb_002170_010 |
0.1682130583 |
- |
- |
hypothetical protein JCGZ_25046 [Jatropha curcas] |
| 17 |
Hb_002066_010 |
0.1690219444 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000201_010 |
0.1698880773 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_002250_140 |
0.170167603 |
- |
- |
PREDICTED: EPIDERMAL PATTERNING FACTOR-like protein 6 isoform X1 [Populus euphratica] |
| 20 |
Hb_000892_030 |
0.1706007934 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |