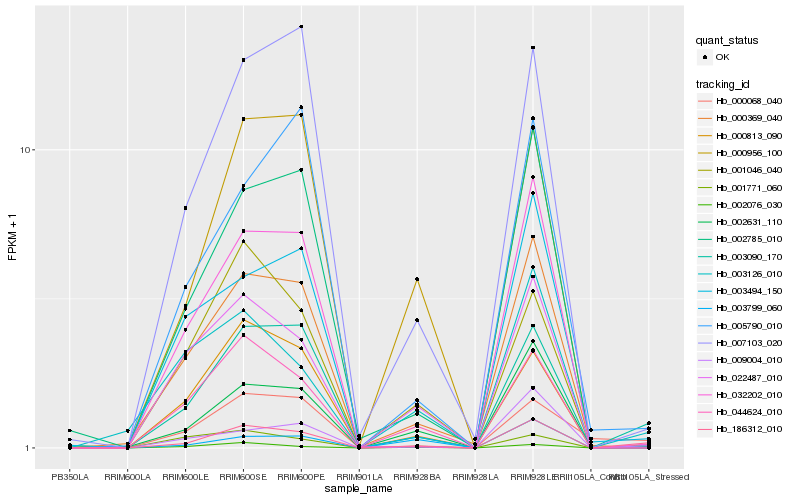
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186312_010 |
0.0 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Populus euphratica] |
| 2 |
Hb_003090_170 |
0.1164139984 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 3 |
Hb_002076_030 |
0.2018885869 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_000369_040 |
0.2021348924 |
- |
- |
Ankyrin repeat family protein, putative [Theobroma cacao] |
| 5 |
Hb_002631_110 |
0.2047075438 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 6 |
Hb_032202_010 |
0.2069288561 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 7 |
Hb_000068_040 |
0.2081414414 |
- |
- |
Inter-alpha-trypsin inhibitor heavy chain H3 precursor, putative [Ricinus communis] |
| 8 |
Hb_003799_060 |
0.210904886 |
- |
- |
hypothetical protein POPTR_0005s22800g [Populus trichocarpa] |
| 9 |
Hb_002785_010 |
0.2114244653 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA-like [Jatropha curcas] |
| 10 |
Hb_007103_020 |
0.2153695474 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 11 |
Hb_001771_060 |
0.2163095753 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 12 |
Hb_044624_010 |
0.2165779404 |
- |
- |
- |
| 13 |
Hb_003126_010 |
0.2177747302 |
- |
- |
PREDICTED: oligopeptide transporter 7-like isoform X1 [Malus domestica] |
| 14 |
Hb_003494_150 |
0.2185801261 |
- |
- |
Silicon transporter, putative [Ricinus communis] |
| 15 |
Hb_009004_010 |
0.2205765654 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At3g14460 [Jatropha curcas] |
| 16 |
Hb_022487_010 |
0.2217159665 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 17 |
Hb_001046_040 |
0.224275291 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 18 |
Hb_000813_090 |
0.2284643651 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 19 |
Hb_000956_100 |
0.229084498 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 20 |
Hb_005790_010 |
0.2304889249 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |