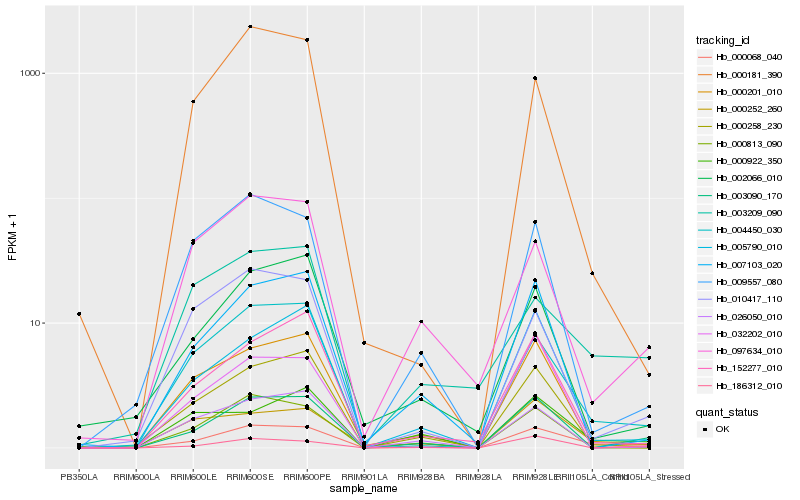
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000068_040 |
0.0 |
- |
- |
Inter-alpha-trypsin inhibitor heavy chain H3 precursor, putative [Ricinus communis] |
| 2 |
Hb_004450_030 |
0.1496834041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003090_170 |
0.1597741495 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 4 |
Hb_000258_230 |
0.1929443137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_005790_010 |
0.2066905365 |
- |
- |
hypothetical protein POPTR_0004s24080g [Populus trichocarpa] |
| 6 |
Hb_186312_010 |
0.2081414414 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Populus euphratica] |
| 7 |
Hb_003209_090 |
0.2084857019 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase family protein [Theobroma cacao] |
| 8 |
Hb_152277_010 |
0.2095888093 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_010417_110 |
0.2112817688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007103_020 |
0.2116353758 |
- |
- |
hypothetical protein POPTR_0007s04191g [Populus trichocarpa] |
| 11 |
Hb_000201_010 |
0.2153467735 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_009557_080 |
0.2169446973 |
- |
- |
PREDICTED: glutaredoxin-C9-like [Populus euphratica] |
| 13 |
Hb_000813_090 |
0.2170034546 |
- |
- |
hypothetical protein CARUB_v10019318mg [Capsella rubella] |
| 14 |
Hb_097634_010 |
0.220466998 |
- |
- |
PREDICTED: UDP-glucuronate 4-epimerase 1 [Jatropha curcas] |
| 15 |
Hb_002066_010 |
0.2208626192 |
- |
- |
PREDICTED: acetylglutamate kinase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000922_350 |
0.2220929807 |
- |
- |
PREDICTED: uncharacterized protein LOC105640367 [Jatropha curcas] |
| 17 |
Hb_000181_390 |
0.2229509618 |
- |
- |
unknown [Lotus japonicus] |
| 18 |
Hb_026050_010 |
0.2230639192 |
- |
- |
protein phosphatase 2c, putative [Ricinus communis] |
| 19 |
Hb_032202_010 |
0.2238377048 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.1 [Jatropha curcas] |
| 20 |
Hb_000252_260 |
0.2242353955 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |