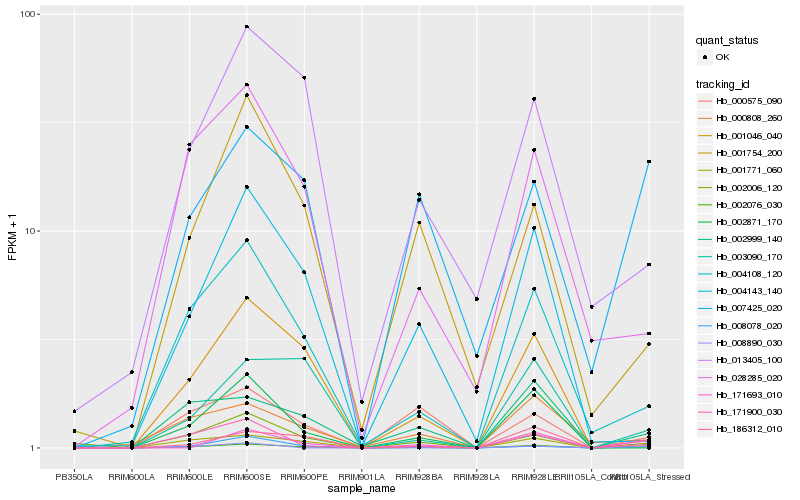
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002076_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 2 |
Hb_171900_030 |
0.1767847489 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 3 |
Hb_001771_060 |
0.1890062383 |
- |
- |
hypothetical protein B456_012G144100 [Gossypium raimondii] |
| 4 |
Hb_004108_120 |
0.1981188327 |
- |
- |
PREDICTED: EID1-like F-box protein 3 [Jatropha curcas] |
| 5 |
Hb_186312_010 |
0.2018885869 |
- |
- |
PREDICTED: O-acyltransferase WSD1-like [Populus euphratica] |
| 6 |
Hb_002999_140 |
0.2187553682 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 7 |
Hb_002006_120 |
0.2204968861 |
- |
- |
PREDICTED: glycine--tRNA ligase 1, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_171693_010 |
0.2285330829 |
- |
- |
hypothetical protein JCGZ_26531 [Jatropha curcas] |
| 9 |
Hb_007425_020 |
0.2347921958 |
transcription factor |
TF Family: ERF |
PREDICTED: pathogenesis-related genes transcriptional activator PTI6-like [Jatropha curcas] |
| 10 |
Hb_008078_020 |
0.2416888492 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |
| 11 |
Hb_000808_260 |
0.2451221551 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 12 |
Hb_028285_020 |
0.2466176128 |
transcription factor |
TF Family: C2C2-CO-like |
zinc finger protein, putative [Ricinus communis] |
| 13 |
Hb_008890_030 |
0.2486026573 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_001754_200 |
0.2486246888 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C [Jatropha curcas] |
| 15 |
Hb_004143_140 |
0.2492339305 |
- |
- |
glycosyltransferase, partial [Arabidopsis thaliana] |
| 16 |
Hb_001046_040 |
0.2510983501 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 17 |
Hb_003090_170 |
0.2563227265 |
- |
- |
PREDICTED: uncharacterized protein LOC105646673 [Jatropha curcas] |
| 18 |
Hb_013405_100 |
0.2566063737 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 8 [Jatropha curcas] |
| 19 |
Hb_000575_090 |
0.2567353326 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 24 [Populus euphratica] |
| 20 |
Hb_002871_170 |
0.25759667 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein 21/22, putative [Ricinus communis] |