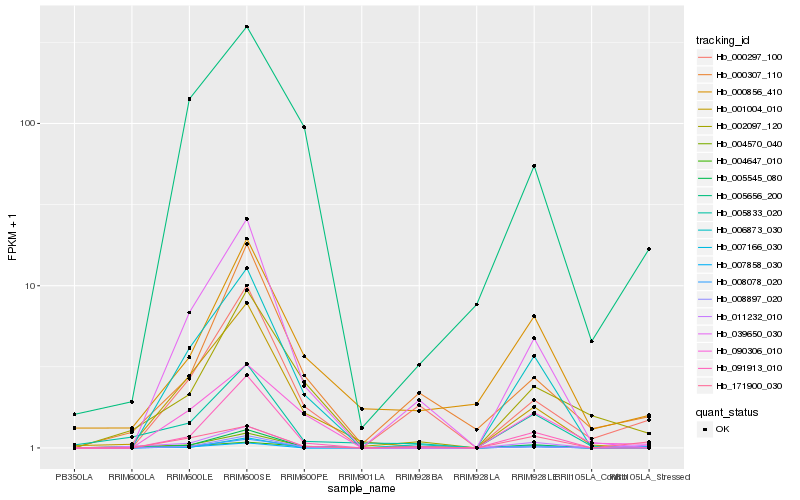
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011232_010 |
0.0 |
- |
- |
Leucine-rich repeat receptor-like protein kinase family protein [Theobroma cacao] |
| 2 |
Hb_008078_020 |
0.2149849032 |
- |
- |
hypothetical protein JCGZ_01297 [Jatropha curcas] |
| 3 |
Hb_004570_040 |
0.2328877668 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 4 |
Hb_171900_030 |
0.2353849843 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein MYBAS2 [Jatropha curcas] |
| 5 |
Hb_006873_030 |
0.2391830355 |
- |
- |
receptor serine/threonine kinase, putative [Ricinus communis] |
| 6 |
Hb_005545_080 |
0.2439712969 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 7 |
Hb_002097_120 |
0.2452573747 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_005833_020 |
0.2483991542 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 [Jatropha curcas] |
| 9 |
Hb_007166_030 |
0.2600969031 |
- |
- |
- |
| 10 |
Hb_001004_010 |
0.2601667949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_091913_010 |
0.2612659711 |
- |
- |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 12 |
Hb_000297_100 |
0.2620838769 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 13 |
Hb_004647_010 |
0.2624546346 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_008897_020 |
0.2655862383 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Musa acuminata subsp. malaccensis] |
| 15 |
Hb_000307_110 |
0.267791109 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 16 |
Hb_005656_200 |
0.2681692947 |
- |
- |
Cation transport protein chaC, putative [Ricinus communis] |
| 17 |
Hb_007858_030 |
0.2696909907 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 18 |
Hb_039650_030 |
0.2697003926 |
- |
- |
hypothetical protein RCOM_2128750 [Ricinus communis] |
| 19 |
Hb_090306_010 |
0.2708219434 |
- |
- |
hypothetical protein POPTR_0007s02300g [Populus trichocarpa] |
| 20 |
Hb_000856_410 |
0.2709224955 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |