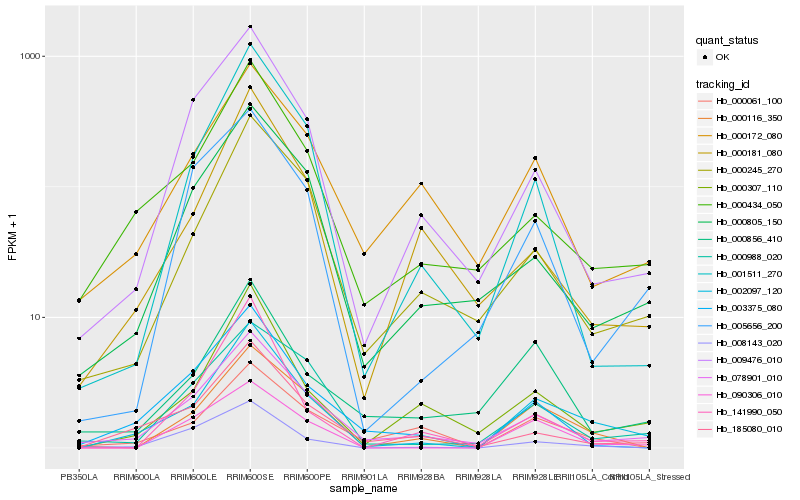
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002097_120 |
0.0 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_078901_010 |
0.1671832369 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 3 |
Hb_000116_350 |
0.1741040951 |
- |
- |
PREDICTED: uncharacterized protein LOC105628582 [Jatropha curcas] |
| 4 |
Hb_000434_050 |
0.181314482 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 5 |
Hb_000307_110 |
0.1838116352 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 6 |
Hb_090306_010 |
0.1896874508 |
- |
- |
hypothetical protein POPTR_0007s02300g [Populus trichocarpa] |
| 7 |
Hb_000245_270 |
0.1931129356 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 8 |
Hb_185080_010 |
0.1947959674 |
- |
- |
- |
| 9 |
Hb_001511_270 |
0.1964064975 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 10 |
Hb_009476_010 |
0.201046365 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 29 [Jatropha curcas] |
| 11 |
Hb_000805_150 |
0.2038400087 |
transcription factor |
TF Family: GRAS |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_000988_020 |
0.2039264295 |
- |
- |
PREDICTED: transcription factor bHLH112 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003375_080 |
0.2071866954 |
- |
- |
PREDICTED: uncharacterized protein LOC105643382 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000061_100 |
0.2083010977 |
- |
- |
hypothetical protein CISIN_1g0053742mg, partial [Citrus sinensis] |
| 15 |
Hb_008143_020 |
0.2108138604 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |
| 16 |
Hb_005656_200 |
0.2146099301 |
- |
- |
Cation transport protein chaC, putative [Ricinus communis] |
| 17 |
Hb_000181_080 |
0.2147478767 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 18 |
Hb_000856_410 |
0.2164418007 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_141990_050 |
0.2191605186 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 20 |
Hb_000172_080 |
0.2196609718 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor RAP2-4-like [Jatropha curcas] |