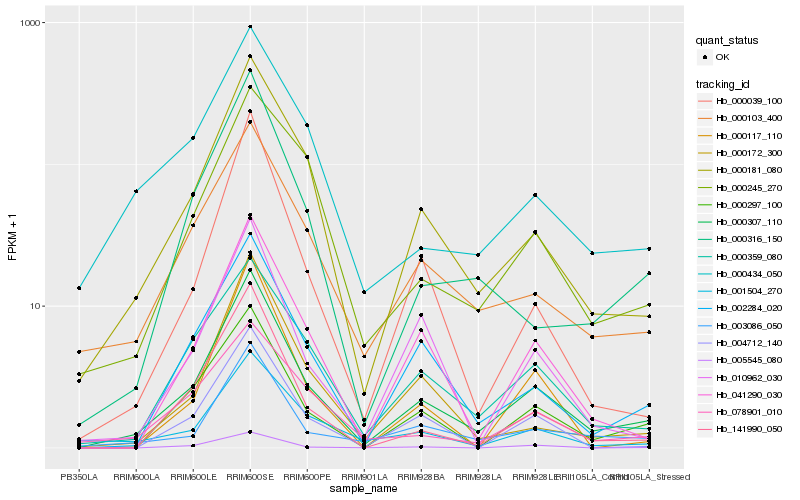
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_110 |
0.0 |
- |
- |
hypothetical protein JCGZ_08160 [Jatropha curcas] |
| 2 |
Hb_000181_080 |
0.113220786 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 3 |
Hb_002284_020 |
0.1349797571 |
- |
- |
PREDICTED: probable inorganic phosphate transporter 1-9 [Jatropha curcas] |
| 4 |
Hb_000297_100 |
0.1394567294 |
transcription factor |
TF Family: NAC |
NAC transcription factor 038 [Jatropha curcas] |
| 5 |
Hb_003086_050 |
0.142615173 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 6 |
Hb_041290_030 |
0.143853636 |
- |
- |
PREDICTED: uncharacterized protein LOC105640874 [Jatropha curcas] |
| 7 |
Hb_000103_400 |
0.1455625754 |
- |
- |
PREDICTED: ATP sulfurylase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000359_080 |
0.146369546 |
- |
- |
basic 7S globulin 2 precursor small subunit, putative [Ricinus communis] |
| 9 |
Hb_141990_050 |
0.1513836032 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 33 [Jatropha curcas] |
| 10 |
Hb_001504_270 |
0.1590149349 |
- |
- |
PREDICTED: pleiotropic drug resistance protein 1-like [Jatropha curcas] |
| 11 |
Hb_000039_100 |
0.1619730337 |
- |
- |
PREDICTED: uncharacterized protein LOC105649440 [Jatropha curcas] |
| 12 |
Hb_004712_140 |
0.1636151279 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 13 |
Hb_000172_300 |
0.1645242967 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 14 |
Hb_000316_150 |
0.1663635732 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 15 |
Hb_078901_010 |
0.1675021682 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 16 |
Hb_000245_270 |
0.1686909883 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 17 |
Hb_000117_110 |
0.1700073381 |
- |
- |
transferase, putative [Ricinus communis] |
| 18 |
Hb_005545_080 |
0.1701172282 |
- |
- |
hypothetical protein JCGZ_01293 [Jatropha curcas] |
| 19 |
Hb_000434_050 |
0.1714850601 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 5 [Hevea brasiliensis] |
| 20 |
Hb_010962_030 |
0.1718123506 |
- |
- |
kinase, putative [Ricinus communis] |